Category:FL
m (→Biosynthesis 生合成) |
(→Biosynthesis 生合成) |
||
| Line 173: | Line 173: | ||
! Explanation (解説) | ! Explanation (解説) | ||
|- | |- | ||
| − | |Flavonoid is synthesized through the phenylpropanoid-acetate pathway in all higher plants. | + | |Flavonoid is synthesized through the phenylpropanoid-acetate pathway in all higher plants (but not algae). Most plants contain the six major subgroups (chalcones, flavones, flavonols, flavandiols, anthycyanins, and proanthocyanidins), but the seventh, aurones, is not ubiquitous. |
| + | |||
| + | |||
| | | | ||
<!----一般にポリフェノール類は | <!----一般にポリフェノール類は | ||
| Line 321: | Line 323: | ||
| chalcone synthase | | chalcone synthase | ||
| Bacterial polyketide synthases, particularly those in fatty acid synthesis (Verwoert et al. 1992) | | Bacterial polyketide synthases, particularly those in fatty acid synthesis (Verwoert et al. 1992) | ||
| − | | | + | | Early response against light <ref>Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM: Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 1992 4:1229-1236</ref> <ref>Pelletier MK, Murrell JR, Shirley BW: Characterization of flavonol synthase and leucoanthocyanidins dioxygenase genes in Arabidopsis. Plant Physiol 1997 113:1437-1445</ref> |
|- | |- | ||
| CHI | | CHI | ||
| chalcone-flavanone isomerase | | chalcone-flavanone isomerase | ||
| − | | Unclear. | + | | Unclear and unique to plants.<ref>Jez JM, Bowman ME, Dixon RA, Noel JP: Structure and mechanism of chalcone isomerase: an evolutionarily unique enzyme in plants. Nat Struct Biol 2000 7: 786–791</ref> |
| − | | | + | ''Eubacterium ramulus'' has the CHI activity. <ref>Herles C, Braune A, Braut M: First bacterial chalcone isomerase isolated from ''Eubacterium ramulus''. Arch Microbiol 2004 181:428-434.</ref> |
| + | | Early response against light. | ||
|- | |- | ||
| F3H | | F3H | ||
| flavanone 3-hydroxylase | | flavanone 3-hydroxylase | ||
| 2-oxoglutarate-dependent dioxygenase family <ref>Winkel-Shirley B: Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology and biotechnology. Plant Physiol 2001 126:485-492</ref> | | 2-oxoglutarate-dependent dioxygenase family <ref>Winkel-Shirley B: Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology and biotechnology. Plant Physiol 2001 126:485-492</ref> | ||
| − | | | + | | Early response in Arabidopsis but late in Antirrhinum <ref>Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E: Control of anthocyanin biosyntehsis in flowers ''Antirrhinum majus''. Plant J 1991 1:37-49</ref> |
|- | |- | ||
| FLS | | FLS | ||
| flavonol synthase | | flavonol synthase | ||
| 2-oxoglutarate-dependent dioxygenase family <ref>Holton TA, Cornish EC: Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1993 7:1071-1083</ref> | | 2-oxoglutarate-dependent dioxygenase family <ref>Holton TA, Cornish EC: Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1993 7:1071-1083</ref> | ||
| − | | | + | | Early response against light. In Arabidopsis, all structural genes are single-copy except for this one, to which six genes exist and two of them are not expressed. |
|- | |- | ||
| DFR | | DFR | ||
| Line 342: | Line 345: | ||
| NADPH-dependent reductase associated with steroid metabolism <ref>Baker ME, Blasco RE: Expansion of the mammalian 3bhydroxysteroid dehydrogenase/plant dihydroflavonol reductase superfamily to include a bacterial cholesterol dehydrogenase, a bacterial UDP-galactose 4-epimerase, and open reading frames in vaccinia virus and fish lymphocystis disease virus. | | NADPH-dependent reductase associated with steroid metabolism <ref>Baker ME, Blasco RE: Expansion of the mammalian 3bhydroxysteroid dehydrogenase/plant dihydroflavonol reductase superfamily to include a bacterial cholesterol dehydrogenase, a bacterial UDP-galactose 4-epimerase, and open reading frames in vaccinia virus and fish lymphocystis disease virus. | ||
FEBS Lett 1992 301: 89–93</ref> | FEBS Lett 1992 301: 89–93</ref> | ||
| − | | | + | | Later response |
|- | |- | ||
| ANS/LDOX | | ANS/LDOX | ||
| anthocyanidin synthase or leucoanthocyanidin dioxygenase | | anthocyanidin synthase or leucoanthocyanidin dioxygenase | ||
| 2-oxoglutarate-dependent dioxygenase family | | 2-oxoglutarate-dependent dioxygenase family | ||
| − | | | + | | Later response |
|- | |- | ||
! colspan=4| Auxiliary Genes (その他の遺伝子) | ! colspan=4| Auxiliary Genes (その他の遺伝子) | ||
| Line 358: | Line 361: | ||
| flavonoid 3',5'-hydroxylase | | flavonoid 3',5'-hydroxylase | ||
| cytochrome P450 hydroxylase family <ref>Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JGT, Lu CY, Farcy E, Stevenson TW, Cornish EC: Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 1993 366:276–279</ref> | | cytochrome P450 hydroxylase family <ref>Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JGT, Lu CY, Farcy E, Stevenson TW, Cornish EC: Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 1993 366:276–279</ref> | ||
| − | | Not reported in mosses or liverworts. | + | | Not reported in mosses or liverworts. The transformation of the F3'5'H and the cytochrome b5 gene of petunia into carnation changed its flower color deep purple.<ref>de Vetten N, ter Horst J, van Schaik H-P, de Boer A, Mol J, Koes R: A cytochrome b5 is required for full activity of flavonoid 39,59-hydroxylase, a cytochrome P450 involved in the formation of blue flowers. Proc Natl Acad Sci USA 1999 96: 778–783</ref><ref>Brugliera F, Tull D, Holton TA, Karan M, Treloar N, |
| + | Simpson K, Skurczynska J, Mason JG: Introduction of a cytochrome b5 enhances the activity of flavonoid 3'5' hydroxylase (a cytochrome P450) in transgenic carnation. Sixth International Congress of Plant Molecular Biology. University of Laval, Quebec, 2000 pp S6–S8</ref> | ||
| + | |- | ||
| + | | FSI | ||
| + | | flavone synthase | ||
| + | | Dioxygenase in parsley (FSI) and P450 monooxygenase in snapdragon (FSII). | ||
| + | |- | ||
| + | | LAR (or LCR) | ||
| + | | leucoanthocyanidin reductase | ||
| + | | | ||
|- | |- | ||
| UF3GT | | UF3GT | ||
| Line 365: | Line 377: | ||
| GST | | GST | ||
| glutathione-S-transferase | | glutathione-S-transferase | ||
| + | | | ||
| + | | Transport of flavonoids from cytoplasm to vacuole or cell walls requires both GST and the glutathione pump in ATP-binding cassette family.<ref>Marrs KA, Alfenito MR, Lloyd AM, Walbot V: A glutathione S-transferase involved in vacuolar transfer encoded by the maise gene Bronze-2. Nature 1995 375: 397–400</ref><ref>Alfenito MR, Souer E, Goodman CD, Buell R, Mol J, Koes R, Walbot V: Functional complementation of anthocyanin sequestration in the vacuole by widely divergent | ||
| + | glutathione S-transferases. Plant Cell 1998 10: 1135–1149</ref> | ||
| + | |- | ||
| + | | AOMT | ||
| + | | anthocyanin O-methyl transferase | ||
|} | |} | ||
Revision as of 21:30, 22 September 2009
Flavonoid (フラボノイド)
| Flavonoid Top | Molecule Index | Author Index | Journals | Structure Search | Food | New Input |
Contents |
Class Overview
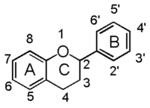
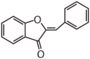
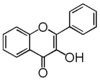
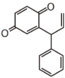
The word "flavonoid" comes from its Latin origin flavus (yellow) with oid, meaning yellow-ish. It comes from its history as yellow natural dye (quercetin and kaempferol are the most widespread flavone dyes. See flavone). Chemically speaking, it is a class of plant secondary metabolites that have two benzene rings (each called A-ring and B-ring) connected by a chain of three carbons (Figure 1).
The carbon chain, corresponding to the numbers 2,3,4 in Figure 1, is linked to a hydroxyl group in the A-ring to form the C-ring. The class of flavonoids are usually determined by the modification pattern of the C-ring (Table 1).
Flavonoid is utilized in many industrial processes from pigments to food additives. Often heard names include anthocyanin, catechin, flavan, and isoflavone.
Biosynthesis 生合成
| Explanation (解説) | |
|---|---|
| Flavonoid is synthesized through the phenylpropanoid-acetate pathway in all higher plants (but not algae). Most plants contain the six major subgroups (chalcones, flavones, flavonols, flavandiols, anthycyanins, and proanthocyanidins), but the seventh, aurones, is not ubiquitous.
|
| 4-coumaroyl CoA + malonyl CoA | |||||||||||||
| | |||||||||||||
| CHALCONES, AURONES | chalcone | FLAVONES | |||||||||||
| |
/FS | ||||||||||||
| IFS / | FLAVANONE naringenin |
kaempferol | FLAVONOLS | quercetin | myricetin | ||||||||
| ISO-FLAVONES | |
/ FLS | / FLS | / FLS | |||||||||
| DIHYDRO FLAVONOLS | dihydro-kaempferol | F3'H |
dihydro-quercetin | F3'5'H |
dihydro-myricetin | ||||||||
| |
|
|
|||||||||||
| LEUCOANTHO-CYANIDINS (flavan diols) |
leuco-pelargonidin | LAR |
leuco-cyanidin | LAR |
leuco-delphinidin | LAR |
|||||||
| |
PROANTHO- CYANIDINS | |
PROANTHO- CYANIDINS | |
PROANTHO- CYANIDINS | ||||||||
| ANTHO-CYANIDINS | pelargonidin | ANR |
|
cyanidin | ANR |
|
delphinidin | ANR |
| ||||
| |
epi-afzelechin | |
epi-catechin | |
epigallo-catechin | ||||||||
| ANTHO-CYANINS | pelargonidin 3-glucoside | cyanidin 3-glucoside | delphinidin 3-glucoside | FLAVAN 3-OLS | |||||||||
|
|
| |||||||||||
| Six Structural Genes (6つの構造遺伝子) | |||
|---|---|---|---|
| Abbrev. | Name | Origin | Information |
| CHS | chalcone synthase | Bacterial polyketide synthases, particularly those in fatty acid synthesis (Verwoert et al. 1992) | Early response against light [1] [2] |
| CHI | chalcone-flavanone isomerase | Unclear and unique to plants.[3]
Eubacterium ramulus has the CHI activity. [4] |
Early response against light. |
| F3H | flavanone 3-hydroxylase | 2-oxoglutarate-dependent dioxygenase family [5] | Early response in Arabidopsis but late in Antirrhinum [6] |
| FLS | flavonol synthase | 2-oxoglutarate-dependent dioxygenase family [7] | Early response against light. In Arabidopsis, all structural genes are single-copy except for this one, to which six genes exist and two of them are not expressed. |
| DFR | dihydroflavonol 4-reductase | NADPH-dependent reductase associated with steroid metabolism [8] | Later response |
| ANS/LDOX | anthocyanidin synthase or leucoanthocyanidin dioxygenase | 2-oxoglutarate-dependent dioxygenase family | Later response |
| Auxiliary Genes (その他の遺伝子) | |||
| F3'H | flavonoid 3'-hydroxylase | cytochrome P450 hydroxylase family [9] | |
| F3'5'H | flavonoid 3',5'-hydroxylase | cytochrome P450 hydroxylase family [10] | Not reported in mosses or liverworts. The transformation of the F3'5'H and the cytochrome b5 gene of petunia into carnation changed its flower color deep purple.[11][12] |
| FSI | flavone synthase | Dioxygenase in parsley (FSI) and P450 monooxygenase in snapdragon (FSII). | |
| LAR (or LCR) | leucoanthocyanidin reductase | ||
| UF3GT | UDP flavonoid glucosyltransferase | ||
| GST | glutathione-S-transferase | Transport of flavonoids from cytoplasm to vacuole or cell walls requires both GST and the glutathione pump in ATP-binding cassette family.[13][14] | |
| AOMT | anthocyanin O-methyl transferase | ||
- ↑ Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM: Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 1992 4:1229-1236
- ↑ Pelletier MK, Murrell JR, Shirley BW: Characterization of flavonol synthase and leucoanthocyanidins dioxygenase genes in Arabidopsis. Plant Physiol 1997 113:1437-1445
- ↑ Jez JM, Bowman ME, Dixon RA, Noel JP: Structure and mechanism of chalcone isomerase: an evolutionarily unique enzyme in plants. Nat Struct Biol 2000 7: 786–791
- ↑ Herles C, Braune A, Braut M: First bacterial chalcone isomerase isolated from Eubacterium ramulus. Arch Microbiol 2004 181:428-434.
- ↑ Winkel-Shirley B: Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology and biotechnology. Plant Physiol 2001 126:485-492
- ↑ Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E: Control of anthocyanin biosyntehsis in flowers Antirrhinum majus. Plant J 1991 1:37-49
- ↑ Holton TA, Cornish EC: Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1993 7:1071-1083
- ↑ Baker ME, Blasco RE: Expansion of the mammalian 3bhydroxysteroid dehydrogenase/plant dihydroflavonol reductase superfamily to include a bacterial cholesterol dehydrogenase, a bacterial UDP-galactose 4-epimerase, and open reading frames in vaccinia virus and fish lymphocystis disease virus. FEBS Lett 1992 301: 89–93
- ↑ Brugliera F, Barri-Rewell G, Holton TA, Mason JG: Isolation and characterization of a flavonoid 3-hydroxylase. cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J 1999 19: 441–451
- ↑ Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JGT, Lu CY, Farcy E, Stevenson TW, Cornish EC: Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 1993 366:276–279
- ↑ de Vetten N, ter Horst J, van Schaik H-P, de Boer A, Mol J, Koes R: A cytochrome b5 is required for full activity of flavonoid 39,59-hydroxylase, a cytochrome P450 involved in the formation of blue flowers. Proc Natl Acad Sci USA 1999 96: 778–783
- ↑ Brugliera F, Tull D, Holton TA, Karan M, Treloar N, Simpson K, Skurczynska J, Mason JG: Introduction of a cytochrome b5 enhances the activity of flavonoid 3'5' hydroxylase (a cytochrome P450) in transgenic carnation. Sixth International Congress of Plant Molecular Biology. University of Laval, Quebec, 2000 pp S6–S8
- ↑ Marrs KA, Alfenito MR, Lloyd AM, Walbot V: A glutathione S-transferase involved in vacuolar transfer encoded by the maise gene Bronze-2. Nature 1995 375: 397–400
- ↑ Alfenito MR, Souer E, Goodman CD, Buell R, Mol J, Koes R, Walbot V: Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 1998 10: 1135–1149
Bioactivity
| all flavonoids
photoprotectant, anti-oxidant |
全フラボノイド
抗紫外線、抗酸化作用 |
| Tannins (proanthocyanidins)
anti-bacteria, anti-fungi |
タンニン (プロアントシアニジニン)
抗菌、抗カビ作用 |
- ↑ 1.0 1.1 Ryan KG, Swinny EE, Markham KR, Winefield C: Flavonoid gene expression and UV photoprotection in transgenic and mutant Penunia leaves. Phytochem 2002 59:23-32
Links to familiar names 耳にする名前
- isoflavonoid in beans (豆のイソフラボン)
- anthocyanin in berries (ベリーのアントシアニン)
- catechin in tea (お茶のカテキン)
- rutin in buckwheat (ソバのルチン)
- hesperidin in orange (ミカンのヘスペリジン)
- naringenin chalcone in tomato (トマトのナリンゲニンカルコン)
Database statistics/ranking データベース統計
This database collects original references that report identification of flavonoid in various plant species. The database consists of three major namespaces: (flavonoid) compounds, plant species, and references. Currently, 6961 flavonoid structures, 3961 plant species, and 5215 references describing total 19861 metabolite-species relationships are registered.
Flavonoid content in food 食品中の量
| Category | Flavonol | Flavone | Flavan | Flavanone |
|---|---|---|---|---|
| Names | quercetin, kampferol, myricetin, isorhamnetin | apigenin, luteolin | catechin, epicatechin | |
| broccoli ブロッコリ | Δ | |||
| celery セロリ | ΔΔ | |||
| fava そら豆 | ΔΔ | |||
| hot pepper とうがらし | ΔΔ | |||
| onion たまねぎ | ΔΔ (Δ) | |||
| parsley パセリ | ΔΔΔ | |||
| peppermint ペパーミント | ΔΔ | |||
| spinach ほうれん草 | Δ | |||
| thyme タイム | ΔΔΔ | |||
| watercress クレソン | Δ | |||
| dill, fennel ディル, フェンネル | ΔΔΔ |
Δ 5 to <10 mg/100 g; ΔΔ 10 to <50 mg/100 g; ΔΔΔ 50< mg/100 g
The following vegetables and herbs have flavonoid contents less than 5 mg/100 g: beets, kidney beans, snap beans, cabbage, carrot, cauliflower, cucumber, endive, gourd, leek, lettuce, green peas, sweet pepper, potato, radish, tomato, oregano, perrilla, rosemary
Design of Flavonoid ID numbers ID番号の設計
12-DIGIT
| F | L | x | x | y | y | z | z | w | c | c | c |
- x ... backbone structure (母核構造)
FL1 aurone and chalcone; FL2 flavanone; FL3 flavone; FL4 Dihydroflavonol; FL5 Flavonol; FL6 Flavan; FL7 Anthocyanin; FLI Isoflavonoid; FLN Neoflavonoid
- y ... hydroxylation pattern in A and B ring (水酸基パターン)
Click above categories to see details. General description is here.
- z ... glycosylation pattern (糖修飾パターン)
Click above categories to see details. General description is here.
- w ... halogenation etc. (ハロゲン等)
Currently unused.
- c ... serial number (通し番号)
For Users of Flavonoid Viewer
The flavonoid IDs used in this site is the same as those in Flavonoid Viewer in metabolome.jp except for the following FL7 category.
| Anthocyanin glycosylated with other than glucose and galactose | ||
|---|---|---|
| Flavonoid Viewer FL7A..GS |
→ | This site FL7A..GO |
Subcategories
This category has the following 10 subcategories, out of 10 total.