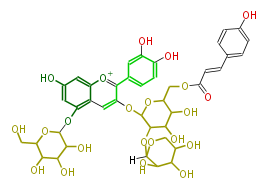
Mol:PR100463
From Metabolomics.JP
ACD/Labs08070915142D 64 70 0 0 1 0 0 0 0 0 2 V2000 13.5444 -7.7957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 13.5444 -8.7571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 14.3770 -9.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 15.2097 -8.7571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 15.2097 -7.7957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 14.3770 -7.3149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 16.0423 -9.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 16.8749 -8.7571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 16.8749 -7.7957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 16.0423 -7.3149 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 21.9816 -11.5364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 22.8142 -11.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 22.8142 -10.0942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 21.9816 -9.6135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 21.1489 -10.0942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 21.1489 -11.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 18.8024 -9.9424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 22.0137 -7.9951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 14.3770 -10.1993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 12.7117 -7.3149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 17.7076 -7.3149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 18.5402 -7.7957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 19.3728 -7.3149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 19.3728 -6.3535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 18.5402 -5.8728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 17.7076 -6.3535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 18.5402 -4.9113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 20.2055 -5.8728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 23.6469 -9.6135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 22.0122 -13.2826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 23.7429 -11.3045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 23.3590 -6.9450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 24.2800 -13.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 24.2800 -14.2774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 25.1126 -14.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 25.9453 -14.2774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 25.9453 -13.3159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 25.1126 -12.8352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 24.2800 -12.3545 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0 25.1126 -11.8737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 26.7779 -14.7581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 26.7779 -12.8352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 23.3590 -5.5830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 24.1916 -5.0061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 25.1370 -5.3426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 25.9696 -4.8619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 22.4141 -5.1023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 26.8023 -5.3426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 27.6349 -4.8619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 27.6349 -3.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 26.8023 -3.4197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 25.9696 -3.9004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 28.4675 -3.4197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 12.4659 -10.3426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 11.6332 -10.8233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 11.6332 -11.7848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 12.4659 -12.2655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 13.2985 -11.7848 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 13.2985 -10.8233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 10.8006 -10.3426 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 12.4659 -13.2269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 11.6332 -13.7077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 12.4659 -9.3812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 10.8006 -12.2655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 1 2 2 0 0 0 0 2 3 1 0 0 0 0 3 4 2 0 0 0 0 4 5 1 0 0 0 0 5 6 2 0 0 0 0 6 1 1 0 0 0 0 4 7 1 0 0 0 0 7 8 2 0 0 0 0 8 9 1 0 0 0 0 9 10 2 0 0 0 0 10 5 1 0 0 0 0 11 12 1 0 0 0 0 12 13 1 0 0 0 0 13 14 1 0 0 0 0 14 15 1 0 0 0 0 15 16 1 0 0 0 0 16 11 1 0 0 0 0 14 18 1 6 0 0 0 3 19 1 0 0 0 0 1 20 1 0 0 0 0 9 21 1 0 0 0 0 21 22 2 0 0 0 0 22 23 1 0 0 0 0 23 24 2 0 0 0 0 24 25 1 0 0 0 0 25 26 2 0 0 0 0 26 21 1 0 0 0 0 25 27 1 0 0 0 0 24 28 1 0 0 0 0 17 8 1 0 0 0 0 13 29 1 1 0 0 0 11 30 1 1 0 0 0 12 31 1 6 0 0 0 18 32 1 0 0 0 0 17 16 1 6 0 0 0 33 34 1 0 0 0 0 34 35 1 0 0 0 0 35 36 1 0 0 0 0 36 37 1 0 0 0 0 37 38 1 0 0 0 0 38 33 1 0 0 0 0 33 39 1 6 0 0 0 38 40 1 6 0 0 0 36 41 1 6 0 0 0 37 42 1 1 0 0 0 33 30 1 0 0 0 0 32 43 1 0 0 0 0 43 44 1 0 0 0 0 44 45 2 0 0 0 0 45 46 1 0 0 0 0 43 47 2 0 0 0 0 46 48 2 0 0 0 0 48 49 1 0 0 0 0 49 50 2 0 0 0 0 50 51 1 0 0 0 0 51 52 2 0 0 0 0 52 46 1 0 0 0 0 50 53 1 0 0 0 0 54 55 1 0 0 0 0 55 56 1 0 0 0 0 56 57 1 0 0 0 0 57 58 1 0 0 0 0 58 59 1 0 0 0 0 59 54 1 0 0 0 0 55 60 1 6 0 0 0 57 61 1 6 0 0 0 61 62 1 0 0 0 0 54 63 1 1 0 0 0 56 64 1 1 0 0 0 19 59 1 6 0 0 0 M CHG 1 10 1 S SKP 7 CAS_RN 763896-30-6 NAME Cyanidin-3-O-(6''-O-(E-p-coum)-2''-O-(beta-xylopyranosyl)-beta-glucopyranoside)-5-O-beta-glucopyranoside ID PR100463 FORMULA C41H45O22 EXACTMASS 889.240248124 AVERAGEMASS 889.7828 SMILES Oc(c1)cc([o+1]6)c(cc(c(c(c7)ccc(c(O)7)O)6)OC(C4OC([H])(O5)C(C(C(C5)O)O)O)OC(C(C4O)O)COC(C=Cc(c3)ccc(O)c3)=O)c1OC(O2)C(O)C(C(C2CO)O)O M END