Mol:FL7AAGGL0052
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
83 90 0 0 0 0 0 0 0 0999 V2000
-1.6660 0.7842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6660 -0.0408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9515 -0.4533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2370 -0.0408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2370 0.7842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9515 1.1967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4774 -0.4533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1919 -0.0408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1919 0.7842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4774 1.1967 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.9685 1.2325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6829 0.8200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3974 1.2325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3974 2.0575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6829 2.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9685 2.0575 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0376 2.4271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0504 -0.5365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2459 1.1190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9515 -1.2160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6829 3.2012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0624 0.8486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3420 -2.5997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4178 -2.9217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9292 -2.2739 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6992 -1.9772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9394 -1.6550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4280 -2.3028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0502 -1.9296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6598 -3.0826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0964 -2.9851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1777 -2.8881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7411 -1.9372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4141 -4.8717 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7087 -4.1023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5223 -3.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1897 -3.4624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4843 -2.6930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2979 -2.5635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8925 -1.6401 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4370 -2.6301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2310 -3.0886 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7110 -3.0756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7110 -3.9666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9453 -4.3837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2352 -3.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5164 -4.3687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5077 -5.1936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2178 -5.6136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9366 -5.2087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2101 -5.5980 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6276 -1.5430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4243 -1.3277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4272 -0.5025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8502 0.2061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0535 -0.0090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0505 -0.8343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4440 -0.8356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9705 -2.1341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0587 -1.6948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0016 -0.8325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0561 4.1866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4680 4.9001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0561 5.6136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2919 4.9001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7039 4.1866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5277 4.1866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9402 4.9011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7652 4.9011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1777 4.1866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7652 3.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9402 3.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0016 4.1866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1635 3.5352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2491 2.8205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0475 3.0296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8409 2.8028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4284 3.5175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6300 3.3085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4730 3.8943 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6467 3.0520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1996 3.0795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8760 2.3897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
8 18 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
15 21 1 0 0 0 0
13 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
28 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
25 32 1 0 0 0 0
26 18 1 0 0 0 0
33 29 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
35 37 1 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
33 38 1 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 45 1 0 0 0 0
48 51 1 0 0 0 0
52 53 1 1 0 0 0
53 54 1 1 0 0 0
55 54 1 1 0 0 0
55 56 1 0 0 0 0
56 57 1 0 0 0 0
57 52 1 0 0 0 0
57 58 1 0 0 0 0
52 59 1 0 0 0 0
53 60 1 0 0 0 0
54 61 1 0 0 0 0
40 58 1 0 0 0 0
62 63 1 0 0 0 0
63 64 2 0 0 0 0
63 65 1 0 0 0 0
65 66 2 0 0 0 0
66 67 1 0 0 0 0
67 68 2 0 0 0 0
68 69 1 0 0 0 0
69 70 2 0 0 0 0
70 71 1 0 0 0 0
71 72 2 0 0 0 0
72 67 1 0 0 0 0
70 73 1 0 0 0 0
74 75 1 1 0 0 0
75 76 1 1 0 0 0
77 76 1 1 0 0 0
77 78 1 0 0 0 0
78 79 1 0 0 0 0
79 74 1 0 0 0 0
79 80 1 0 0 0 0
74 81 1 0 0 0 0
75 82 1 0 0 0 0
76 83 1 0 0 0 0
62 80 1 0 0 0 0
21 77 1 0 0 0 0
55 22 1 0 0 0 0
S SKP 8
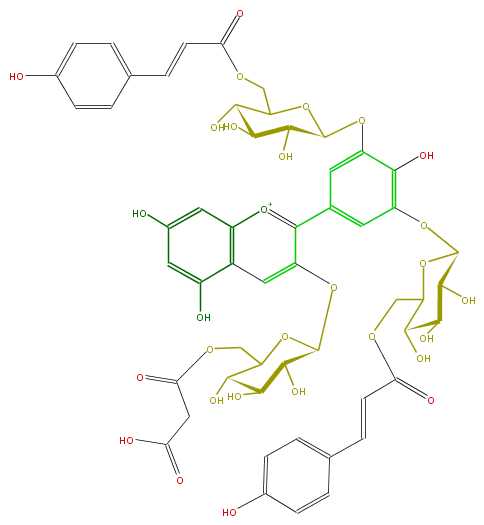
ID FL7AAGGL0052
KNApSAcK_ID C00014803
NAME Delphinidin 3-(6-malonylglucoside)-3',5'-di-(6-p-coumaroylglucoside);Ternatin D3
CAS_RN 215378-79-3
FORMULA C54H55O29
EXACTMASS 1167.282900798
AVERAGEMASS 1167.9971
SMILES c(c7)(OC(C(O)8)OC(COC(CC(O)=O)=O)C(O)C8O)c([o+1]c(c76)cc(cc6O)O)c(c3)cc(c(O)c3OC(O4)C(C(O)C(C4COC(=O)C=Cc(c5)ccc(O)c5)O)O)OC(O1)C(C(O)C(C1COC(=O)C=Cc(c2)ccc(O)c2)O)O
M END