Mol:FL7AACGL0101
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
112122 0 0 0 0 0 0 0 0999 V2000
-2.5754 2.2573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9744 2.6043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9744 3.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5754 3.6453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1764 3.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1764 2.6043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3734 2.2573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7724 2.6043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7724 3.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3734 3.6453 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
-0.1366 3.6654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4319 3.3372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0004 3.6654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0004 4.3218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4319 4.6500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1366 4.3218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5072 4.6144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7374 3.6222 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5754 1.6303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1912 2.2688 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4319 5.3146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0201 0.7534 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-4.2935 0.3626 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.7246 0.9601 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.9306 1.1842 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.6572 1.5752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2262 0.9777 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.6595 1.4110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1838 1.4110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3133 0.2456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1992 -0.1726 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3770 0.6125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3589 -0.2262 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.5634 -0.0074 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.5643 0.8176 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.1446 1.5279 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.9401 1.3092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9393 0.4842 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-1.0129 -0.8255 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8058 -0.4273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1210 1.0013 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2329 1.6167 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.8204 0.9023 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6136 1.1290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4117 0.9198 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8243 1.6342 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0310 1.4076 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.3000 1.8737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7611 1.4903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0590 1.9694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9250 0.7330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0272 0.1535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9649 -0.5584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6682 -0.4344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6546 -1.0015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9215 -1.5737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5534 -2.0993 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9191 -2.0438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5538 -2.5654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8229 -3.1426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4573 -3.1981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8226 -2.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5219 -3.5725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7007 -3.7200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0997 -3.5608 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.3520 -3.9094 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.1318 -4.1787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6656 -4.8078 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
5.4133 -4.4593 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.6335 -4.1899 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
4.3887 -2.9411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1411 -3.9531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.3795 -4.7170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0788 -5.7184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4968 -6.5316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6079 0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9402 -0.2706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9402 -0.9121 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6781 -0.7144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9402 -1.5389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3516 -1.8788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3516 -2.4784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9312 -2.8130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9312 -3.4823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3516 -3.8169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7720 -3.4823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7720 -2.8130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3516 -4.3603 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3250 -3.7403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3111 -4.2960 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.8986 -5.0105 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.1005 -4.8014 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3073 -5.0282 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.7197 -4.3137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5179 -4.5227 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.8697 -4.3945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1797 -5.4975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8042 -5.3146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8819 -3.8923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4413 -3.7424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8315 -3.0666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3803 -2.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4126 -3.0666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7629 -3.6732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.5340 -3.6732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.8625 -3.1043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.5194 -3.1043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.8478 -3.6732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.5194 -4.2421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.8625 -4.2421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-8.4315 -3.6732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-7.9667 -2.6569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 3 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
5 18 1 0 0 0 0
1 19 1 0 0 0 0
8 20 1 0 0 0 0
15 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
19 25 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
22 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
20 36 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 41 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
41 43 1 0 0 0 0
47 48 1 0 0 0 0
42 49 1 0 0 0 0
46 50 1 0 0 0 0
45 51 1 0 0 0 0
51 52 1 0 0 0 0
52 53 1 0 0 0 0
53 54 2 0 0 0 0
53 55 1 0 0 0 0
55 56 2 0 0 0 0
56 57 1 0 0 0 0
57 58 2 0 0 0 0
58 59 1 0 0 0 0
59 60 2 0 0 0 0
60 61 1 0 0 0 0
61 62 2 0 0 0 0
62 57 1 0 0 0 0
60 63 1 0 0 0 0
61 64 1 0 0 0 0
65 66 1 0 0 0 0
66 67 1 1 0 0 0
68 67 1 1 0 0 0
69 68 1 1 0 0 0
69 70 1 0 0 0 0
70 65 1 0 0 0 0
64 66 1 0 0 0 0
65 71 1 0 0 0 0
70 72 1 0 0 0 0
69 73 1 0 0 0 0
68 74 1 0 0 0 0
74 75 1 0 0 0 0
38 76 1 0 0 0 0
76 77 1 0 0 0 0
77 78 1 0 0 0 0
78 79 2 0 0 0 0
78 80 1 0 0 0 0
80 81 2 0 0 0 0
81 82 1 0 0 0 0
82 83 2 0 0 0 0
83 84 1 0 0 0 0
84 85 2 0 0 0 0
85 86 1 0 0 0 0
86 87 2 0 0 0 0
87 82 1 0 0 0 0
85 88 1 0 0 0 0
86 89 1 0 0 0 0
90 91 1 1 0 0 0
91 92 1 1 0 0 0
93 92 1 1 0 0 0
93 94 1 0 0 0 0
94 95 1 0 0 0 0
95 90 1 0 0 0 0
88 93 1 0 0 0 0
90 96 1 0 0 0 0
91 97 1 0 0 0 0
92 98 1 0 0 0 0
95 99 1 0 0 0 0
99100 1 0 0 0 0
100101 1 0 0 0 0
101102 2 0 0 0 0
101103 1 0 0 0 0
103104 2 0 0 0 0
104105 1 0 0 0 0
105106 2 0 0 0 0
106107 1 0 0 0 0
107108 2 0 0 0 0
108109 1 0 0 0 0
109110 2 0 0 0 0
110105 1 0 0 0 0
108111 1 0 0 0 0
107112 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 74 75
M SBL 2 1 81
M SMT 2 CH2OH
M SVB 2 81 8.1008 -2.3298
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 28 29
M SBL 1 1 31
M SMT 1 CH2OH
M SVB 1 31 -4.6595 1.411
S SKP 8
ID FL7AACGL0101
KNApSAcK_ID C00011110
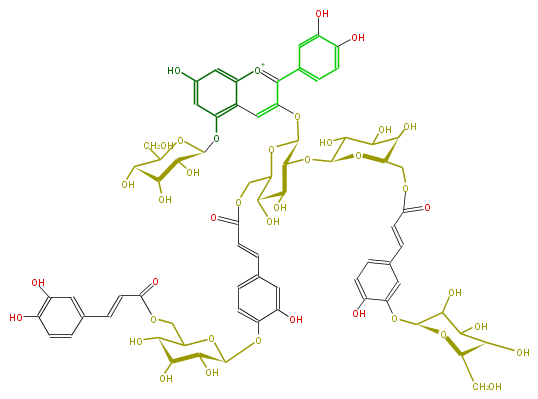
NAME Cyanidin 3-O-[2-O-(6-O-(trans-3-O-(beta-D-glucopyranosyl)caffeyl)-beta-D-glucopyranosyl)-6-O-(trans-4-O-(6-O-(trans-caffeyl)-beta-D-glucopyranposyl)caffeyl)-beta-D-glucopyranoside]-5-O-[beta-D-glucopyranoside]
CAS_RN 756795-04-7
FORMULA C72H79O40
EXACTMASS 1583.414762408
AVERAGEMASS 1584.37366
SMILES c(O)(c(O)%11)cc(cc%11)c(c1O[C@@H]([C@H]4O[C@H](O8)C(O)C(O)[C@@H](O)[C@@H](COC(C=Cc(c%10)cc(c(O)c%10)O[C@H](O9)C(O)C([C@H]([C@@H](CO)9)O)O)=O)8)OC(COC(=O)C=Cc(c7)cc(c(c7)O[C@H](O5)[C@@H](O)[C@H]([C@H](C5COC(C=Cc(c6)ccc(O)c(O)6)=O)O)O)O)[C@@H]([C@@H]4O)O)[o+1]c(c2)c(c(O[C@H](O3)[C@H]([C@H]([C@@H](O)C3CO)O)O)cc2O)c1
M END