Mol:FL7AACGL0077
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
82 89 0 0 0 0 0 0 0 0999 V2000
-3.1887 1.1835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1887 0.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6324 0.2200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0761 0.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0761 1.1835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6324 1.5047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5198 0.2200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9635 0.5412 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9635 1.1835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5198 1.5047 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
-0.4074 1.5046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1596 1.1773 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7266 1.5046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7266 2.1593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1596 2.4866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4074 2.1593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7448 1.5046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2934 2.4865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6324 -0.4221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5474 -0.1795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5517 0.1171 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
0.9534 -0.4132 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.5319 -0.1882 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0900 -0.1822 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6845 0.2235 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.1784 -0.0436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3272 -0.4437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8633 -0.7446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0812 -0.3143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1596 3.1411 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7912 -0.6963 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-4.4200 -1.1863 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.8855 -0.9784 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.3697 -0.9729 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.7445 -0.5980 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2121 -0.8448 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-5.3049 -0.3997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7703 -1.6475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5792 -1.4925 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5827 -0.2696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2440 0.1341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6845 0.2235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5011 0.8655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0368 1.1423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6372 0.7956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0368 1.7820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4686 2.0313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4686 2.7111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0297 3.0350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0297 3.6829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4686 4.0068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9076 3.6829 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9076 3.0350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4686 4.6544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3418 -2.6291 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
0.2403 -2.4789 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.2126 -1.8842 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.5032 -1.3710 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.1353 -1.5963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0789 -2.2160 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.3418 -3.1020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7459 -1.8769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1797 -2.7206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8567 -3.2872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7202 -2.7206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0190 -3.2809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6168 -3.2809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9247 -2.7406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5208 -2.7406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8088 -3.2809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5009 -3.8211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9049 -3.8211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4042 -3.2809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0214 -3.4555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6865 -1.8031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5868 -2.2384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4950 -4.2419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2094 -4.6544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5905 4.0067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3050 3.5942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9979 -2.1078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7124 -2.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
22 27 1 0 0 0 0
23 28 1 0 0 0 0
24 29 1 0 0 0 0
15 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
36 40 1 0 0 0 0
40 41 1 0 0 0 0
34 19 1 0 0 0 0
25 42 1 0 0 0 0
42 43 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
44 43 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 52 1 0 0 0 0
52 53 2 0 0 0 0
53 48 1 0 0 0 0
51 54 1 0 0 0 0
55 56 1 1 0 0 0
56 57 1 1 0 0 0
58 57 1 1 0 0 0
58 59 1 0 0 0 0
59 60 1 0 0 0 0
60 55 1 0 0 0 0
55 61 1 0 0 0 0
57 62 1 0 0 0 0
63 64 2 0 0 0 0
63 65 1 0 0 0 0
65 66 2 0 0 0 0
66 67 1 0 0 0 0
67 68 2 0 0 0 0
68 69 1 0 0 0 0
69 70 2 0 0 0 0
70 71 1 0 0 0 0
71 72 2 0 0 0 0
72 67 1 0 0 0 0
70 73 1 0 0 0 0
20 21 1 0 0 0 0
27 58 1 0 0 0 0
62 63 1 0 0 0 0
56 74 1 0 0 0 0
60 75 1 0 0 0 0
75 76 1 0 0 0 0
71 77 1 0 0 0 0
77 78 1 0 0 0 0
50 79 1 0 0 0 0
79 80 1 0 0 0 0
69 81 1 0 0 0 0
81 82 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 75 76
M SBL 5 1 82
M SMT 5 CH2OH
M SVB 5 82 -0.6865 -1.8031
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 40 41
M SBL 4 1 42
M SMT 4 CH2OH
M SVB 4 42 -4.677 -0.3427
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 81 82
M SBL 3 1 88
M SMT 3 OCH3
M SVB 3 88 3.9979 -2.1078
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 79 80
M SBL 2 1 86
M SMT 2 OCH3
M SVB 2 86 4.5905 4.0067
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 77 78
M SBL 1 1 84
M SMT 1 OCH3
M SVB 1 84 3.495 -4.2419
S SKP 8
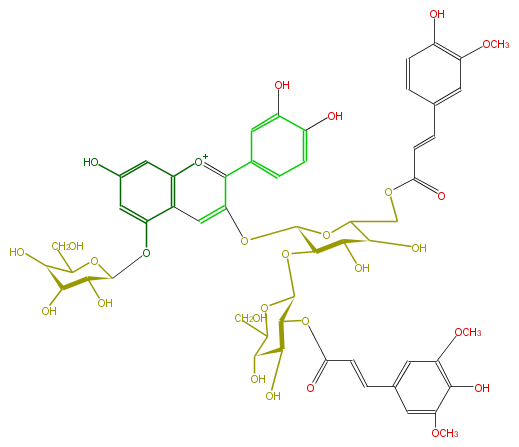
ID FL7AACGL0077
KNApSAcK_ID C00006853
NAME Cyanidin 3-(6''-ferulyl-2'''-sinapylsophoroside)-5-glucoside
CAS_RN 117894-46-9,124088-76-2,110202-92-1
FORMULA C54H59O28
EXACTMASS 1155.3192863040001
AVERAGEMASS 1156.0294600000002
SMILES Oc(c8)c(cc(c8)c(c3O[C@H]([C@@H]5O[C@H](O6)[C@@H](OC(=O)C=Cc(c7)cc(c(c(OC)7)O)OC)[C@H]([C@@H](O)C6CO)O)OC([C@H]([C@@H](O)5)O)COC(=O)C=Cc(c4)cc(c(c4)O)OC)[o+1]c(c2)c(c3)c(cc2O)O[C@@H]([C@H]1O)OC(CO)[C@@H]([C@@H]1O)O)O
M END