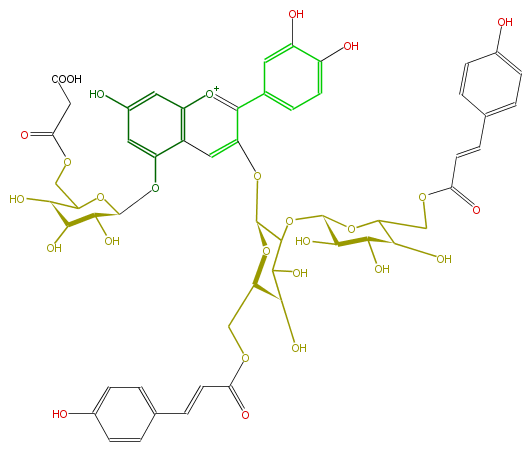
Mol:FL7AACGL0076
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
82 89 0 0 0 0 0 0 0 0999 V2000
-2.6129 2.1286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6129 1.4863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0566 1.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5003 1.4863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5003 2.1286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0566 2.4498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9440 1.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3877 1.4863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3877 2.1286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9440 2.4498 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.1684 2.4497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7354 2.1224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3023 2.4497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3023 3.1044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7354 3.4317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1684 3.1044 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1690 2.4497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8692 3.4316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0566 0.5230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0284 0.7656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5069 -0.5485 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.0001 -0.1609 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.2094 -0.7589 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0437 -1.4569 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.5069 -1.7749 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.3322 -1.1635 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.6816 -0.1437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8337 -1.2074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8114 -2.7274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5684 -2.3082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3514 -0.0462 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.6549 -0.6381 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.2635 -0.5171 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8143 -0.6081 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4853 -0.1381 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.9405 -0.3133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0328 -0.5594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4934 -1.1226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7675 -0.9104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7354 4.0862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2059 -2.9423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2154 0.2488 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.8442 -0.2412 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.3097 -0.0333 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.7939 -0.0278 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.1687 0.3471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6363 0.1003 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.8063 0.3005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1946 -0.7024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0034 -0.5474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1012 0.6024 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8377 1.0588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5438 -3.5276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2161 -4.0952 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1106 -3.5276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4347 -4.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0615 -4.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3740 -3.5476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9990 -3.5476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3115 -4.0889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9990 -4.6301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3740 -4.6301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9358 -4.0889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4701 -0.3118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4010 0.3524 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9766 0.5319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5077 0.0863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0877 1.1619 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5564 1.3325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6744 2.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2831 2.2235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3956 2.8615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8994 3.2779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2906 3.0563 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1781 2.4184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0118 3.9156 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1590 1.6154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7200 1.6154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8007 2.2360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1026 2.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7002 2.7589 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7580 3.3558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
27 31 1 0 0 0 0
22 20 1 0 0 0 0
15 40 1 0 0 0 0
30 41 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
47 51 1 0 0 0 0
51 52 1 0 0 0 0
45 19 1 0 0 0 0
41 53 1 0 0 0 0
53 54 2 0 0 0 0
53 55 1 0 0 0 0
55 56 2 0 0 0 0
56 57 1 0 0 0 0
57 58 2 0 0 0 0
58 59 1 0 0 0 0
59 60 2 0 0 0 0
60 61 1 0 0 0 0
61 62 2 0 0 0 0
62 57 1 0 0 0 0
60 63 1 0 0 0 0
35 64 1 0 0 0 0
64 65 1 0 0 0 0
66 67 2 0 0 0 0
66 68 1 0 0 0 0
68 69 2 0 0 0 0
69 70 1 0 0 0 0
66 65 1 0 0 0 0
70 71 2 0 0 0 0
71 72 1 0 0 0 0
72 73 2 0 0 0 0
73 74 1 0 0 0 0
74 75 2 0 0 0 0
75 70 1 0 0 0 0
73 76 1 0 0 0 0
52 77 1 0 0 0 0
77 78 2 0 0 0 0
77 79 1 0 0 0 0
79 80 1 0 0 0 0
80 81 1 0 0 0 0
80 82 2 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 80 82 81
M SBL 1 1 87
M SMT 1 COOH
M SVB 1 87 -4.1026 2.7589
S SKP 8
ID FL7AACGL0076
KNApSAcK_ID C00006852
NAME Cyanidin 3-(6'',6'''-di-p-coumarylsophoroside)-5-(6-malonylglucoside)
CAS_RN 176260-03-0
FORMULA C54H55O28
EXACTMASS 1151.287986176
AVERAGEMASS 1151.9977
SMILES [C@@H]([C@H]8O)([C@@H](OC(COC(=O)CC(O)=O)[C@H](O)8)Oc(c1)c(c3)c([o+1]c(c(O[C@@H](C(O[C@@H](O6)[C@H](O)[C@@H]([C@@H](C(COC(=O)C=Cc(c7)ccc(c7)O)6)O)O)5)O[C@@H]([C@H](O)C5O)COC(=O)C=Cc(c4)ccc(O)c4)3)c(c2)cc(c(c2)O)O)cc1O)O
M END