Mol:FL5FACGS0058
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
65 71 0 0 0 0 0 0 0 0999 V2000
-3.5971 0.4737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7086 -0.1589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2165 -0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6129 -0.3521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5014 0.2805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9934 0.6934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1208 -0.7650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5172 -0.5453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4057 0.0873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8977 0.5002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2077 -1.2582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6387 0.4037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1371 -0.0171 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4781 0.2068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5918 0.8516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0902 1.2724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5250 1.0485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3279 -1.2041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2502 1.0912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0261 1.0101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2039 1.9169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9605 -1.0125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8713 -1.5926 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.2730 -2.1229 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8515 -1.8979 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.4097 -1.8919 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.0041 -1.4862 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.4980 -1.7533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6468 -2.1534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1829 -2.4543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0041 -1.4862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4009 -2.0240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4934 -0.8768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9625 -0.6348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4760 -0.9313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9625 -0.1311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2403 2.0185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7378 1.5969 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.8058 2.2492 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.5182 2.8426 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.0206 3.2643 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.9527 2.6119 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.9760 3.7747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1337 2.9826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4364 1.9393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2434 -2.0341 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
0.2302 -2.4315 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.1661 -1.8166 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.4372 -1.2572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0364 -0.8598 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.0277 -1.4746 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.6741 -1.5887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6787 -2.3389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2302 -2.9305 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6336 -2.2091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5254 0.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5254 0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1324 1.2268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1324 1.9277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5254 2.2782 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9183 1.9277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9183 1.2268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5254 2.9780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5807 2.9443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4277 2.4128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
16 21 1 0 0 0 0
22 8 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
24 29 1 0 0 0 0
25 30 1 0 0 0 0
27 31 1 0 0 0 0
26 32 1 0 0 0 0
31 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
34 36 1 0 0 0 0
38 37 1 1 0 0 0
38 39 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
41 42 1 1 0 0 0
42 37 1 1 0 0 0
41 43 1 0 0 0 0
40 44 1 0 0 0 0
39 45 1 0 0 0 0
20 38 1 0 0 0 0
47 46 1 1 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
49 50 1 0 0 0 0
50 51 1 1 0 0 0
51 46 1 1 0 0 0
51 52 1 0 0 0 0
46 53 1 0 0 0 0
47 54 1 0 0 0 0
48 55 1 0 0 0 0
23 52 1 0 0 0 0
50 22 1 0 0 0 0
36 56 2 0 0 0 0
56 57 1 0 0 0 0
57 58 2 0 0 0 0
58 59 1 0 0 0 0
59 60 2 0 0 0 0
60 61 1 0 0 0 0
61 62 2 0 0 0 0
62 57 1 0 0 0 0
60 63 1 0 0 0 0
42 64 1 0 0 0 0
64 65 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 64 65
M SBL 1 1 70
M SMT 1 CH2OH
M SVB 1 70 -5.1324 2.2283
S SKP 8
ID FL5FACGS0058
KNApSAcK_ID C00006000
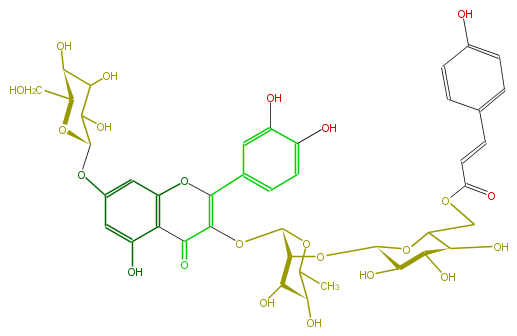
NAME Quercetin 3-(6'''-p-coumarylglucosyl)(1->2)-rhamnoside-7-glucoside
CAS_RN 143016-73-3
FORMULA C42H46O23
EXACTMASS 918.242987778
AVERAGEMASS 918.8008399999999
SMILES c(c1)(ccc(C=CC(OCC(O2)[C@@H](O)[C@H]([C@@H](O)[C@H]2O[C@H]([C@@H](OC(C5=O)=C(Oc(c6)c(c(O)cc6O[C@H](C(O)7)O[C@@H](CO)[C@H](C7O)O)5)c(c4)cc(c(O)c4)O)3)[C@@H]([C@H](O)C(C)O3)O)O)=O)c1)O
M END