Mol:FL5FACGL0074
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
67 73 0 0 0 0 0 0 0 0999 V2000
-2.7577 1.0510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7577 0.4087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2014 0.0875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6451 0.4087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6451 1.0510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2014 1.3722 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0888 0.0875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5325 0.4087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5325 1.0510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0888 1.3722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0888 -0.4133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0236 1.3721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5905 1.0448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1575 1.3721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1575 2.0268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5905 2.3541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0236 2.0268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3138 1.3721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7243 2.3540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1384 0.0198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2014 -0.5546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5905 3.0086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8802 -1.9779 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.2981 -1.8276 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.3259 -1.2330 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0352 -0.7197 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.6737 -0.9451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6173 -1.5648 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.8802 -2.4507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2981 -2.5840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2075 -1.2257 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3074 -1.8466 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.6686 -1.8466 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.1278 -2.2908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2847 -2.9138 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.9236 -2.9138 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.4644 -2.4696 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.9229 -2.2558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2889 -3.0841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2202 -0.7545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8171 -1.1576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9342 -0.2590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7453 -0.7545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9723 -0.3612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4260 -0.3612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6988 -0.8337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2444 -0.8337 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5172 -0.3612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2444 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6988 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0617 -0.3612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5166 -1.3052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7212 2.1926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1533 1.8647 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.3335 2.4952 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.1533 3.1296 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.7212 3.4575 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.5411 2.8269 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.7659 3.9678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7989 3.3342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9159 2.2541 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4593 -3.4073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1738 -3.8198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2250 -1.1519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1253 -1.5872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2173 3.0452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9592 2.3746 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 20 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
36 39 1 0 0 0 0
33 31 1 0 0 0 0
40 41 1 0 0 0 0
41 32 1 0 0 0 0
40 42 2 0 0 0 0
40 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 45 1 0 0 0 0
48 51 1 0 0 0 0
47 52 1 0 0 0 0
54 53 1 1 0 0 0
54 55 1 0 0 0 0
55 56 1 0 0 0 0
56 57 1 0 0 0 0
57 58 1 1 0 0 0
58 53 1 1 0 0 0
57 59 1 0 0 0 0
56 60 1 0 0 0 0
55 61 1 0 0 0 0
18 54 1 0 0 0 0
35 62 1 0 0 0 0
62 63 1 0 0 0 0
28 64 1 0 0 0 0
64 65 1 0 0 0 0
58 66 1 0 0 0 0
66 67 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 66 67
M SBL 3 1 72
M SMT 3 CH2OH
M SVB 3 72 -4.0617 3.3105
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 64 65
M SBL 2 1 70
M SMT 2 CH2OH
M SVB 2 70 -1.225 -1.1519
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 62 63
M SBL 1 1 68
M SMT 1 CH2OH
M SVB 1 68 1.4593 -3.4073
S SKP 8
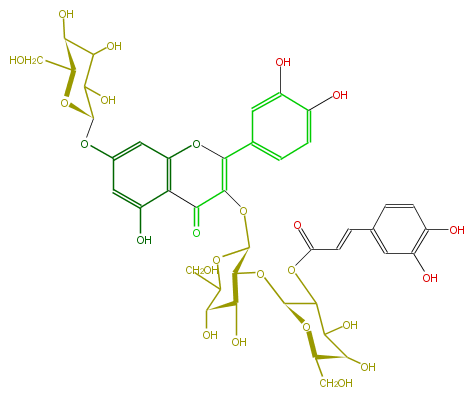
ID FL5FACGL0074
KNApSAcK_ID C00005996
NAME Quercetin 3-(2'''-(E)-caffeylsophoroside]-7-glucoside
CAS_RN 151649-67-1
FORMULA C42H46O25
EXACTMASS 950.2328170220001
AVERAGEMASS 950.79964
SMILES [C@@H](CO)(O1)[C@H](O)C(C(OC(=O)C=Cc(c7)ccc(O)c7O)[C@H](O[C@@H]([C@@H](O)6)[C@@H](OC(CO)[C@@H]6O)OC(C2=O)=C(c(c5)cc(O)c(O)c5)Oc(c3)c(c(cc3O[C@@H](O4)C(C([C@H](O)[C@@H]4CO)O)O)O)2)1)O
M END