Mol:FL5FACGL0066
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
66 72 0 0 0 0 0 0 0 0999 V2000
-3.8175 1.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8175 0.7912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2612 0.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7049 0.7912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7049 1.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2612 1.7547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1486 0.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5923 0.7912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5923 1.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1486 1.7547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1486 -0.0308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0362 1.7546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4692 1.4273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0978 1.7546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0978 2.4093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4692 2.7366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0362 2.4093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3736 1.7546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6646 2.7366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1981 0.4023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2612 -0.1721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4692 3.3911 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9400 -1.5954 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.3578 -1.4451 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.3856 -0.8504 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.0950 -0.3372 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.7334 -0.5626 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6771 -1.1822 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-1.9400 -2.0682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3578 -2.2015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8523 -0.8432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2477 -1.4641 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.3912 -1.4641 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.0680 -1.9083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2250 -2.5313 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.8638 -2.5313 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.4047 -2.0871 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.8631 -1.8733 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2291 -2.7016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8546 0.5546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1009 0.9602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6718 0.9413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8655 1.4649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1154 1.8895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8488 2.4612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1802 2.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9360 3.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3604 3.5085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0290 3.0352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2732 2.5115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1164 4.0319 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5211 -0.3342 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
0.9228 -0.8644 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.5012 -0.6395 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0181 -0.7159 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6538 -0.2277 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.1477 -0.4948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2426 -0.7751 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2966 -0.8949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8327 -1.1959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3763 -0.9228 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1748 0.1642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2847 -0.7694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1850 -1.2048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3996 -3.0248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1141 -3.4373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 20 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
36 39 1 0 0 0 0
33 31 1 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 45 1 0 0 0 0
48 51 1 0 0 0 0
52 53 1 1 0 0 0
53 54 1 1 0 0 0
55 54 1 1 0 0 0
55 56 1 0 0 0 0
56 57 1 0 0 0 0
57 52 1 0 0 0 0
52 58 1 0 0 0 0
53 59 1 0 0 0 0
54 60 1 0 0 0 0
58 32 1 0 0 0 0
55 61 1 0 0 0 0
56 62 1 0 0 0 0
62 40 1 0 0 0 0
28 63 1 0 0 0 0
63 64 1 0 0 0 0
35 65 1 0 0 0 0
65 66 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 65 66
M SBL 2 1 71
M SMT 2 CH2OH
M SVB 2 71 0.3996 -3.0248
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 63 64
M SBL 1 1 69
M SMT 1 CH2OH
M SVB 1 69 -2.2847 -0.7694
S SKP 8
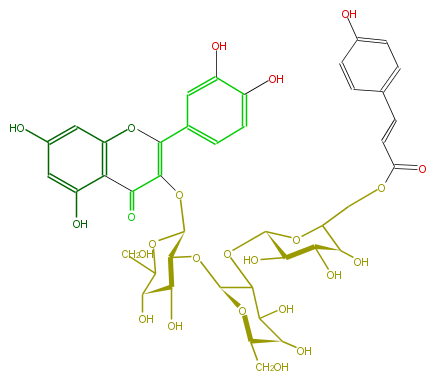
ID FL5FACGL0066
KNApSAcK_ID C00005985
NAME Quercetin 3-(6''''-p-coumarylsophorotrioside)
CAS_RN 167638-65-5
FORMULA C42H46O24
EXACTMASS 934.2379024
AVERAGEMASS 934.8002399999999
SMILES O[C@H](C(O)5)[C@H](O[C@H](C5O[C@H]([C@@H]6O)OC(COC(=O)C=Cc(c7)ccc(O)c7)[C@H]([C@H]6O)O)O[C@@H]([C@H]1O)[C@H](OC(C(=O)2)=C(c(c4)cc(c(c4)O)O)Oc(c3)c2c(O)cc(O)3)OC([C@H](O)1)CO)CO
M END