Mol:FL5FAAGL0102
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
69 75 0 0 0 0 0 0 0 0999 V2000
-1.3296 1.8280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3296 1.1857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7733 0.8645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2170 1.1857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2170 1.8280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7733 2.1492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3393 0.8645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8956 1.1857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8956 1.8280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3393 2.1492 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3393 0.3637 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5961 2.2729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1630 1.9455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7300 2.2729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7300 2.9275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1630 3.2549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5961 2.9275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7733 0.2224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3368 3.2779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8452 2.2818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6281 0.5954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7965 1.9939 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.4524 1.5397 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.9569 1.7324 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.4787 1.7376 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.8262 2.0851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3323 1.9033 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.1811 2.2160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9888 1.5136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4981 1.4205 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9907 -0.5359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9535 0.1205 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.3843 0.0922 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2134 0.5957 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9100 0.9649 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3995 0.9224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5857 0.4179 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-2.1450 -0.2495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6658 0.5064 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3077 0.8647 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.7094 0.3345 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2878 0.5594 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.8460 0.5654 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.4404 0.9711 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.9343 0.7040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0832 0.3040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6193 0.0030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0320 0.2998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0832 -0.5256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5881 -0.8171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4651 -0.8532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4922 -1.4514 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9357 -1.7727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3949 -1.4604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1459 -1.7727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1459 -2.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3949 -2.7093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9357 -2.3971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6865 -2.7092 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9340 0.6226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6816 0.2741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5113 2.5530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4790 2.8050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5471 1.2880 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2229 1.7611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5031 -1.1539 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0030 -0.2878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0493 -2.8654 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6652 -3.2779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
31 32 1 0 0 0 0
35 30 1 0 0 0 0
25 20 1 0 0 0 0
40 41 1 1 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 40 1 0 0 0 0
41 46 1 0 0 0 0
42 47 1 0 0 0 0
43 48 1 0 0 0 0
21 40 1 0 0 0 0
46 49 1 0 0 0 0
49 50 2 0 0 0 0
49 51 1 0 0 0 0
51 52 2 0 0 0 0
52 53 1 0 0 0 0
53 54 2 0 0 0 0
54 55 1 0 0 0 0
55 56 2 0 0 0 0
56 57 1 0 0 0 0
57 58 2 0 0 0 0
58 53 1 0 0 0 0
56 59 1 0 0 0 0
37 60 1 0 0 0 0
60 61 1 0 0 0 0
27 62 1 0 0 0 0
62 63 1 0 0 0 0
44 64 1 0 0 0 0
64 65 1 0 0 0 0
55 66 1 0 0 0 0
66 67 1 0 0 0 0
57 68 1 0 0 0 0
68 69 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 64 65
M SBL 5 1 70
M SMT 5 CH2OH
M SVB 5 70 3.7741 0.95
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 62 63
M SBL 4 1 68
M SMT 4 CH2OH
M SVB 4 68 -3.5113 2.553
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 60 61
M SBL 3 1 66
M SMT 3 CH2OH
M SVB 3 66 -3.5645 1.0767
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 68 69
M SBL 2 1 74
M SMT 2 OCH3
M SVB 2 74 -0.0493 -2.8654
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 66 67
M SBL 1 1 72
M SMT 1 OCH3
M SVB 1 72 -0.5031 -1.1539
S SKP 8
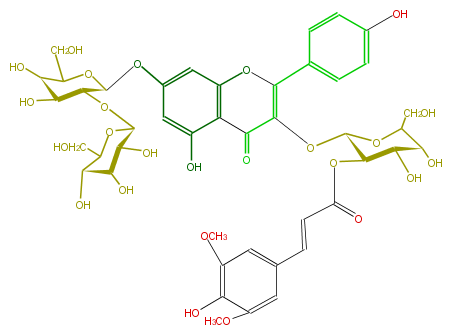
ID FL5FAAGL0102
KNApSAcK_ID C00005922
NAME Kaempferol 3-(2''-sinapylglucoside)-7-sophoroside
CAS_RN 112208-89-6
FORMULA C44H50O25
EXACTMASS 978.26411715
AVERAGEMASS 978.8528000000001
SMILES C(=Cc(c7)cc(OC)c(c(OC)7)O)C(O[C@H]([C@@H]1O)[C@@H](OC(C(=O)5)=C(c(c6)ccc(O)c6)Oc(c54)cc(cc(O)4)O[C@@H]([C@H]2O[C@@H]([C@H]3O)OC([C@@H]([C@H](O)3)O)CO)OC(CO)[C@@H]([C@@H]2O)O)OC([C@@H](O)1)CO)=O
M END