Mol:FL5FAAGL0101
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
66 72 0 0 0 0 0 0 0 0999 V2000
0.0559 1.0923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0559 0.2626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7745 -0.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4930 0.2626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4930 1.0923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7745 1.5071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2115 -0.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9303 0.2626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9303 1.0923 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2115 1.5071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2115 -0.7992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6484 1.5070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3807 1.0842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1132 1.5070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1132 2.3526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3807 2.7755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6484 2.3526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7217 1.4823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.8453 2.7754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4407 -0.3033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7745 -0.9816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0300 -1.4479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3155 -1.0353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5421 -1.8286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3155 -2.6269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0300 -3.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8034 -2.2461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9724 -0.7671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2685 -2.5548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0240 3.5095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3963 2.7615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7570 2.3354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3840 1.6427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1524 2.4671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6670 2.8910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5445 3.8108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6994 3.3723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7635 1.4128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1240 0.7604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3543 0.4543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7724 -2.1778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4639 -2.5770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2445 -1.8092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4639 -1.0368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7724 -0.6374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9917 -1.4053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8397 -1.4053 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8543 -2.8009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3397 -3.2111 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.8044 -2.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5776 0.3068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3817 -0.4245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9670 -1.0098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6632 -0.8231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1731 -1.3330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9865 -2.0293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2901 -2.2158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7804 -1.7062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4951 -2.5379 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8472 -3.4537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6534 3.6304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7308 3.0976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9188 -0.6087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.8453 0.2912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3994 -3.8108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6458 -3.0573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
23 20 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
18 33 1 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
38 32 1 0 0 0 0
42 41 1 1 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 1 0 0 0
46 41 1 1 0 0 0
46 47 1 0 0 0 0
41 48 1 0 0 0 0
42 49 1 0 0 0 0
43 50 1 0 0 0 0
45 28 1 0 0 0 0
39 51 1 0 0 0 0
51 52 2 0 0 0 0
52 53 1 0 0 0 0
53 54 2 0 0 0 0
54 55 1 0 0 0 0
55 56 2 0 0 0 0
56 57 1 0 0 0 0
57 58 2 0 0 0 0
58 53 1 0 0 0 0
56 59 1 0 0 0 0
26 60 1 0 0 0 0
61 62 1 0 0 0 0
35 61 1 0 0 0 0
63 64 1 0 0 0 0
55 63 1 0 0 0 0
65 66 1 0 0 0 0
25 65 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 61 62
M SBL 1 1 68
M SMT 1 CH2OH
M SBV 1 68 -0.0135 -0.7395
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 63 64
M SBL 2 1 70
M SMT 2 ^ OCH3
M SBV 2 70 0.7457 -0.7242
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 65 66
M SBL 3 1 72
M SMT 3 CH2OH
M SBV 3 72 -0.0839 1.1840
S SKP 5
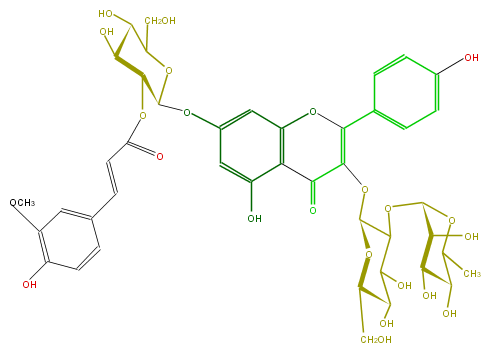
ID FL5FAAGL0101
FORMULA C43H48O23
EXACTMASS 932.258637842
AVERAGEMASS 932.8274200000001
SMILES C(O2)(=C(OC(C(OC(C(O)7)OC(C)C(C(O)7)O)6)OC(C(C6O)O)CO)C(=O)c(c(O)5)c2cc(c5)OC(O3)C(OC(C=Cc(c4)cc(c(O)c4)OC)=O)C(O)C(O)C3CO)c(c1)ccc(O)c1
M END