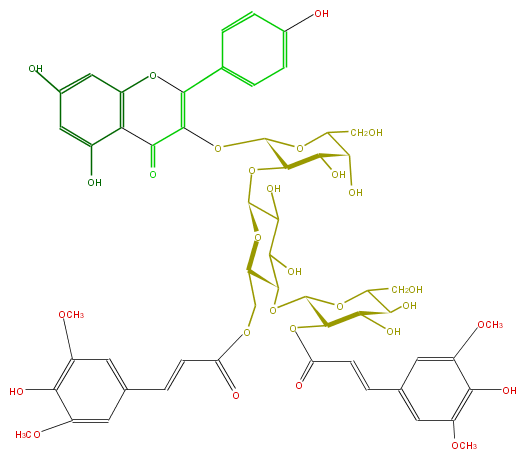
Mol:FL5FAAGA0038
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
84 91 0 0 0 0 0 0 0 0999 V2000
-4.8874 3.4326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8874 2.6046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1702 2.1905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4531 2.6046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4531 3.4326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1702 3.8466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7360 2.1905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0189 2.6046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0189 3.4326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7360 3.8466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7360 1.5449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1159 4.0060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3852 3.5840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3457 4.0060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3457 4.8500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3852 5.2719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1159 4.8500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1702 1.3628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1279 5.3015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.5520 4.0175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2076 2.1361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1189 2.3503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3990 1.6668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1446 1.9568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8641 1.9645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3413 2.4875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6889 2.1432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4084 1.6274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5257 1.5354 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9015 1.1271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2074 0.4521 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5058 0.8638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2795 0.0719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5058 -0.7247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2074 -1.1364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0188 -0.3446 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0024 1.2167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4854 -0.7339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0743 -1.6396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3382 -1.5374 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5471 -2.1419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2344 -2.8292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7849 -3.6076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0310 -2.8292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4260 -3.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3611 -3.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7727 -2.8004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5960 -2.8004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0076 -3.5132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5960 -4.2262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7727 -4.2262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8300 -3.5132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8164 -1.3306 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3342 -2.0142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0798 -1.7241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7993 -1.7164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2765 -1.1934 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6241 -1.5377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5268 -2.0535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7972 -2.1334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1613 -1.5074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9861 -2.8488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6724 -3.3924 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9043 -2.8488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2824 -3.5039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0375 -3.5039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4499 -4.2182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2749 -4.2182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6873 -3.5039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2749 -2.7895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4499 -2.7895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5115 -3.5039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4377 -4.5220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5167 -5.0538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9584 2.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8793 1.9891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3175 -4.7698 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2385 -5.3015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8936 -1.1395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8146 -1.6712 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8457 -1.7178 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9247 -2.2495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9090 -1.9607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.8300 -2.4924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
23 28 1 0 0 0 0
24 29 1 0 0 0 0
25 30 1 0 0 0 0
21 22 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
34 40 1 0 0 0 0
32 28 1 0 0 0 0
40 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
44 45 2 0 0 0 0
45 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 46 1 0 0 0 0
49 52 1 0 0 0 0
53 54 1 1 0 0 0
54 55 1 1 0 0 0
56 55 1 1 0 0 0
56 57 1 0 0 0 0
57 58 1 0 0 0 0
58 53 1 0 0 0 0
54 59 1 0 0 0 0
55 60 1 0 0 0 0
56 61 1 0 0 0 0
39 53 1 0 0 0 0
59 62 1 0 0 0 0
62 63 2 0 0 0 0
62 64 1 0 0 0 0
64 65 2 0 0 0 0
65 66 1 0 0 0 0
66 67 2 0 0 0 0
67 68 1 0 0 0 0
68 69 2 0 0 0 0
69 70 1 0 0 0 0
70 71 2 0 0 0 0
71 66 1 0 0 0 0
69 72 1 0 0 0 0
73 74 1 0 0 0 0
50 73 1 0 0 0 0
75 76 1 0 0 0 0
26 75 1 0 0 0 0
77 78 1 0 0 0 0
68 77 1 0 0 0 0
79 80 1 0 0 0 0
57 79 1 0 0 0 0
81 82 1 0 0 0 0
48 81 1 0 0 0 0
83 84 1 0 0 0 0
70 83 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 73 74
M SBL 1 1 81
M SMT 1 ^OCH3
M SBV 1 81 0.8416 0.2958
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 75 76
M SBL 2 1 83
M SMT 2 CH2OH
M SBV 2 83 -0.6172 -0.0333
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 77 78
M SBL 3 1 85
M SMT 3 OCH3
M SBV 3 85 -0.0425 0.5516
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 79 80
M SBL 4 1 87
M SMT 4 CH2OH
M SBV 4 87 -0.6172 -0.0539
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 81 82
M SBL 5 1 89
M SMT 5 ^OCH3
M SBV 5 89 0.2497 -1.0826
M STY 1 6 SUP
M SLB 1 6 6
M SAL 6 2 83 84
M SBL 6 1 91
M SMT 6 OCH3
M SBV 6 91 -0.6341 -0.8289
S SKP 5
ID FL5FAAGA0038
FORMULA C55H60O29
EXACTMASS 1184.322025958
AVERAGEMASS 1185.0475
SMILES O(C(C(O)5)C(OC(=C7c(c8)ccc(O)c8)C(=O)c(c6O)c(O7)cc(c6)O)OC(CO)C5O)C(O1)C(C(C(OC(C3OC(=O)C=Cc(c4)cc(OC)c(c4OC)O)OC(CO)C(C3O)O)C1COC(C=Cc(c2)cc(OC)c(c(OC)2)O)=O)O)O
M END