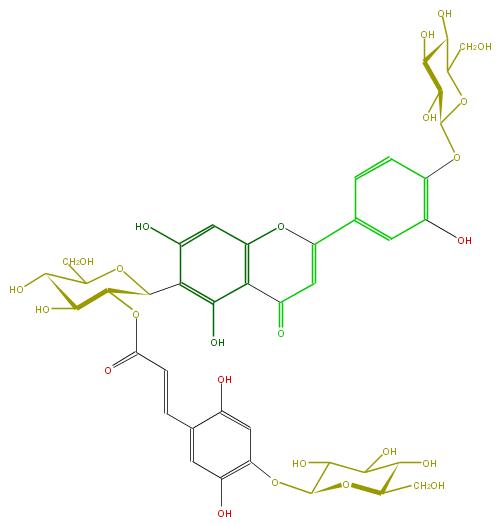
Mol:FL3FACDS0020
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
67 73 0 0 0 0 0 0 0 0999 V2000
-1.8283 0.3821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8283 -0.4744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0866 -0.9027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3449 -0.4744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3449 0.3821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0866 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3969 -0.9027 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1386 -0.4744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1386 0.3821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3969 0.8104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3969 -1.5704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0866 -1.7588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0450 0.9477 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8266 0.4964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6083 0.9477 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6083 1.8502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8266 2.3014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0450 1.8502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3096 2.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8203 -0.2642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1329 -1.0342 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4453 -0.6216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5103 -0.7316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1703 -0.1267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9129 -0.4842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3868 -0.5912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8012 -1.0342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5914 0.8227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8060 -1.1299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3894 0.4967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0979 4.9653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5765 4.4440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9416 3.8035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9416 3.0664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4629 3.5876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0979 4.2281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9427 5.5443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5765 5.0740 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6079 3.2254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8060 -2.0143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4252 -2.3718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1459 -2.3954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1459 -3.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5705 -3.6904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5705 -4.4326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9279 -4.8036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2852 -4.4326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2852 -3.6904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9279 -3.3194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9279 -2.5787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9279 -5.5443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2602 -4.8566 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4835 -4.4384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0824 -5.1330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8536 -4.9125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6296 -5.1330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0308 -4.4384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2595 -4.6586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8950 -4.4384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7313 -4.1869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6221 -4.4384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3835 0.0309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1613 0.8086 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4341 4.8106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3868 4.2606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4064 -4.9249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3591 -5.4749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
2 23 1 0 0 0 0
1 28 1 0 0 0 0
22 29 1 0 0 0 0
15 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
34 19 1 0 0 0 0
33 39 1 0 0 0 0
29 40 1 0 0 0 0
40 41 2 0 0 0 0
40 42 1 0 0 0 0
42 43 2 0 0 0 0
43 44 1 0 0 0 0
44 45 2 0 0 0 0
45 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 44 1 0 0 0 0
49 50 1 0 0 0 0
46 51 1 0 0 0 0
47 52 1 0 0 0 0
53 54 1 0 0 0 0
54 55 1 1 0 0 0
55 56 1 1 0 0 0
57 56 1 1 0 0 0
57 58 1 0 0 0 0
58 53 1 0 0 0 0
53 59 1 0 0 0 0
58 60 1 0 0 0 0
57 61 1 0 0 0 0
54 52 1 0 0 0 0
62 63 1 0 0 0 0
25 62 1 0 0 0 0
64 65 1 0 0 0 0
36 64 1 0 0 0 0
66 67 1 0 0 0 0
56 66 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 62 63
M SBL 1 1 69
M SMT 1 ^CH2OH
M SBV 1 69 0.4707 -0.5151
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 64 65
M SBL 2 1 71
M SMT 2 CH2OH
M SBV 2 71 -0.3363 -0.5824
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 66 67
M SBL 3 1 73
M SMT 3 CH2OH
M SBV 3 73 -0.7768 -0.2081
S SKP 5
ID FL3FACDS0020
FORMULA C42H46O25
EXACTMASS 950.2328170220001
AVERAGEMASS 950.79964
SMILES c(c45)(c(c(O)cc4OC(c(c7)cc(c(c7)OC(O6)C(C(O)C(O)C6CO)O)O)=CC5=O)C(O1)C(OC(=O)C=Cc(c2O)cc(c(OC(O3)C(C(O)C(C3CO)O)O)c2)O)C(C(C(CO)1)O)O)O
M END