Koryoginsenoside-R2
From Metabolomics.JP
Upper classes
| IDs and Links | |
|---|---|
| LipidBank | [1] |
| LipidMaps | [2] |
| CAS | 171746-13-7 |
| KEGG | {{{KEGG}}} |
| KNApSAcK | |
| CDX file | |
| MOL file | Koryoginsenoside-R2.mol |
| Koryoginsenoside R2 | |
|---|---|
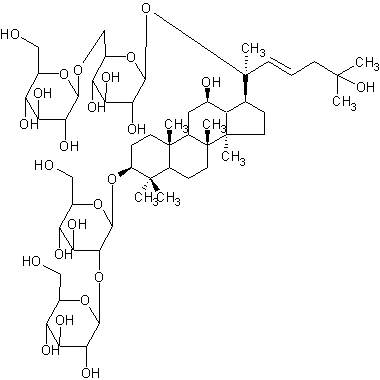
| |
| Structural Information | |
| Systematic Name | (3beta,12beta,22E)-20-((6-O-beta-D-Glucopyranosyl-beta-D-glucopyranosyl)oxy)-12,25-dihydroxydammar-22-en-3-yl 2-O-beta-D-glucopyranosyl-beta-D-glucopyranoside |
| Common Name |
|
| Symbol | |
| Formula | C54H92O24 |
| Exact Mass | 1124.597853872 |
| Average Mass | 1125.29388 |
| SMILES | O(C1OC(C8(C)C)CCC(C78)(C(C(C)(CC7)3)CC(C(C(C(C)(OC |
| Physicochemical Information | |
| Melting Point | |
| Boiling Point | |
| Density | |
| Optical Rotation | |
| Reflactive Index | |
| Solubility | |
| Spectral Information | |
| Mass Spectra | |
| UV Spectra | |
| IR Spectra | |
| NMR Spectra | |
| Chromatograms | |
Spectroscopic Data
| M.P. | 177 - 180 °C |
| 1H-NMR (C5D5N, 400 MHz) | 0.83 (s, 3xH-30), 0.88 (s, 3xH-19), 1.02 (s, 3xH-18), 1.09 (s, 3xH-29), 1.27 (s, 3xH-28), 1.58 (s, 3xH-27), 1.62 (s, 3xH-26), 1.63 (s, 3xH-21), 2.87 (dd, J=13.2, 5.8 Hz, H-22alpha), 3.09 (dd, J=12.5, 4.7 Hz, H-22beta), 3.25 (dd, J=11.5, 4.2 Hz, H-3), 4.02 (dd, J=10.7, 8.2 Hz, H-12), 4.90 (d, J=7.6 Hz, H-1 of Glc II), 5.09 (d, J=7.5 Hz, H-1 of Glc III), 5.17 (d, J=7.8 Hz, H-1 of Glc IV), 5.37 (d, J=7.8 Hz, H-1 of Glc I), 6.12 (d, J=15.6 Hz, H-24), 6.15 (ddd, J=15.6, 8.5 Hz, H-23) |
| 13C-NMR (C5D5N, 100 MHz) | C-1) 39.3, (2) 26.8, (3) 89.2, (4) 40.2, (5) 56.6, (6) 18.6, (7) 35.2, (8) 40.2, (9) 50.3, (10) 37.1, (11) 31.0, (12) 70.6, (13) 49.7, (14) 51.6, (15) 30.7, (16) 26.4, (17) 52.2, (18) 16.3, (19) 16.1, (20) 83.5, (21) 23.5, (22) 138.2, (23) 126.7, (24) 39.8, (25) 81.4, (26) 25.5, (27) 25.2, (28) 28.2, (29) 16.7, (30) 17.3 Glc I (1) 105.0, (2) 83.3, (3) 78.3, (4) 71.8, (5) 78.4, (6) 63.0 Glc II (1) 106.0, (2) 77.1, (3) 78.8, (4) 71.9, (5) 78.4, (6) 63.0 Glc III (1) 98.2, (2) 75.3, (3) 78.0, (4) 71.9, (5) 77.1, (6) 70.1 Glc IV (1) 105.1, (2) 75.3, (3) 78.3, (4) 71.9, (5) 78.4, (6) 63.0 |
C.-R. Yang et al., Phytochemistry, 40, 4193 (1995).