Category:FL/Biosynthesis
m (→{{Bilingual|生合成|Biosynthesis}}) |
m |
||
| Line 1: | Line 1: | ||
| − | =={{Bilingual| | + | =={{Bilingual|フラボノイドの生合成|Biosynthesis of Flavonoid}}== |
{{Twocolumn | {{Twocolumn | ||
| Line 242: | Line 242: | ||
同様に、カロテノイドの生合成遺伝子において下流の酵素はより選択圧が低いことが示されています。 | 同様に、カロテノイドの生合成遺伝子において下流の酵素はより選択圧が低いことが示されています。 | ||
}} | }} | ||
| + | |||
| + | |||
| + | <references/> | ||
Latest revision as of 15:35, 3 July 2014
[edit] Biosynthesis of Flavonoid
Flavonoid is synthesized through both the phenylpropanoid-acetate pathway and the acetate-malonate pathway in all higher plants (but not algae). Most plants contain the 6 major subgroups: chalcones, flavanones, flavones, flavonols, flavans, and anthocyani(di)ns. Aurones or isoflavonoids are not ubiquitous.
| 4-coumaroyl CoA + malonyl CoA | ||||||||||||||||
| | ||||||||||||||||
| CHALCONES, AURONES |
chalconaringenin |
FLAVONES |
apigenin |
luteolin | ||||||||||||
| CHI |
| |||||||||||||||
IFS |
naringenin FLAVANONE |
kaempferol |
FLAVONOLS |
quercetin |
myricetin | |||||||||||
| ISO-FLAVONES |
F3H |
|||||||||||||||
| DIHYDRO FLAVONOLS |
dihydro-kaempferol |
F3'H +OH in B-ring |
dihydro-quercetin |
F3'5'H +OH in B-ring |
dihydro-myricetin | |||||||||||
| DFR |
|
|
||||||||||||||
| LEUCOANTHO-CYANIDINS |
leuco-pelargonidin |
|
leuco-cyanidin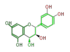
|
leuco-delphinidin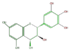
|
||||||||||||
| LDOX |
PROANTHO- CYANIDINS * | |
PROANTHO- CYANIDINS * | |
*...polymers of flavanols | |||||||||||
| ANTHO-CYANIDINS |
pelargonidin |
|
cyanidin |
|
delphinidin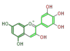
|
| ||||||||||
| UF3GT |
epi-afzelechin |
|
epi-catechin |
|
epigallo-catechin |
FLAVAN 3-OLS | ||||||||||
| ANTHO-CYANINS |
pelargonidin 3-glucoside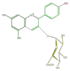
|
cyanidin 3-glucoside |
delphinidin 3-glucoside | |||||||||||||
|
|
| ||||||||||||||
| Information for the Above Abbreviated Gene Names | |||
|---|---|---|---|
| Six Structural Genes | |||
| Abbrev. | Name | Origin | Information |
| CHS | chalcone synthase | Bacterial polyketide synthases, particularly those in fatty acid synthesis (Verwoert et al. 1992) | Early response against light [1] [2] |
| CHI | chalcone-flavanone isomerase | Unclear and unique to plants.[3]
Eubacterium ramulus has the CHI activity. [4] |
Early response against light. |
| F3H | flavanone 3-hydroxylase | 2-oxoglutarate-dependent dioxygenase family [5] | Early response in Arabidopsis but late in Antirrhinum [6] |
| FLS | flavonol synthase | 2-oxoglutarate-dependent dioxygenase family [7] | Early response against light. In Arabidopsis, all structural genes are single-copy except for this one, to which six genes exist and two of them are not expressed. |
| DFR | dihydroflavonol 4-reductase | NADPH-dependent reductase associated with steroid metabolism [8] | Later response |
| ANS/LDOX | anthocyanidin synthase or leucoanthocyanidin dioxygenase | 2-oxoglutarate-dependent dioxygenase family | Later response |
| Auxiliary Genes | |||
| F3'H | flavonoid 3'-hydroxylase | cytochrome P450 hydroxylase family [9] | |
| F3'5'H | flavonoid 3',5'-hydroxylase | cytochrome P450 hydroxylase family [10] | Not reported in mosses or liverworts. The transformation of the F3'5'H and the cytochrome b5 gene of petunia into carnation changed its flower color deep purple.[11][12] |
| FSI | flavone synthase | Dioxygenase in parsley (FSI) and P450 monooxygenase in snapdragon (FSII). | |
| LAR (or LCR) | leucoanthocyanidin reductase | ||
| UF3GT | UDP flavonoid glucosyltransferase | ||
| GST | glutathione-S-transferase | Transport of flavonoids from cytoplasm to vacuole or cell walls requires both GST and the glutathione pump in ATP-binding cassette family.[13][14] | |
| AOMT | anthocyanin O-methyl transferase | ||
| |||
[edit] Evolutionary Pressure
Rausher and colleagues studied the relationship between pathway architecture and protein evolutionary rates in anthocyanin biosynthetic pathways. Upstream enzymes showed reduced rates of non-synonymous substitution compared with downstream enzymes, which are under relaxed constraints, in both broad (Zea in monocot and Antirrhinum and Ipomoea in eudicot [1] [2] ) and narrow (within Ipomoea [3]) comparisons. Similarly, the downstream enzyme is under less selective pressure in the carotenoid biosynthetic pathway [4].
- ↑ Rausher MD, Miller R, Tiffin P (1999) "Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway" Mol Biol Evol 16:266-274
- ↑ Lu Y, Rausher MD (2003) "Evolutionary rate variation in anthocyanin pathway genes" Mol Biol Evol 20:1844-1853
- ↑ Rausher MD, Lu Y, Meyer K (2008) "Variation in constraint versus positive selection as an explanation for evolutionary rate variation among anthocyanin genes" J Mol Evol 7:137-144
- ↑ Livingstone K, Anderson S (2009) "Patterns of variation in the evolution of carotenoid biosynthetic pathway enzymes of higher plants" J Heredity 100:754-761
This category currently contains no pages or media.