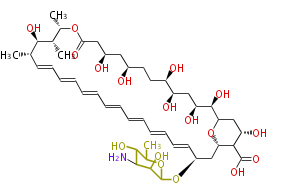
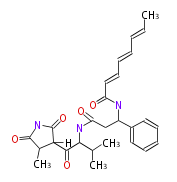
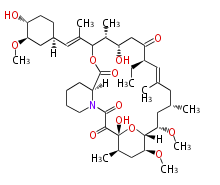
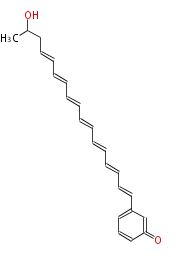
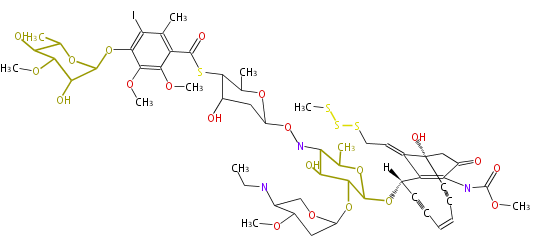
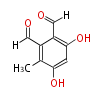
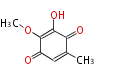
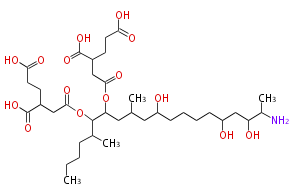
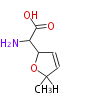
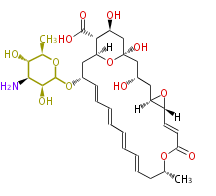
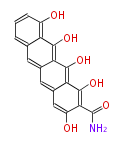
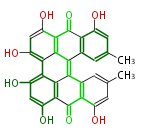
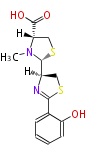
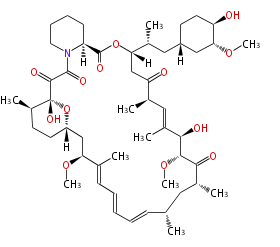
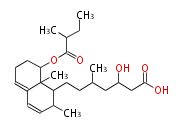Category:PK
m |
m (→Polyketide Synthase (PKS)) |
||
| Line 251: | Line 251: | ||
==Polyketide Synthase (PKS) == | ==Polyketide Synthase (PKS) == | ||
| − | ===Distribution=== | + | ==={{Bilingual|分布|Distribution}}=== |
| + | {{Twocolumn| | ||
PKS members are found in bacteria, fungi, plants, slime mold<ref>Zucko J, Skunca N, Curk T, Zupan B, Long PF et al (2007) "Polyketide synthase genes and the natural products potential of Dictyostelium discoideum" ''Bioinformatics'' 23:2543-49</ref>, Alveolata<ref>Zhu G, LaGier MJ, Stejskal F, Millership JJ, Cai X et al (2002) "Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase" ''Gene'' 298:79-89</ref>, and animals <ref>Castoe TA, Stephens T, Noonan BP, Calestani C (2007) "A novel group of type I polyketide synthases (PKS) in animals and the complex phylogenomics of PKSs" ''Gene'' 392:47-58</ref><ref>Calestani C, Rast JP, Davidson EH (2003) "Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening" ''Development'' 130:4587-96</ref>. | PKS members are found in bacteria, fungi, plants, slime mold<ref>Zucko J, Skunca N, Curk T, Zupan B, Long PF et al (2007) "Polyketide synthase genes and the natural products potential of Dictyostelium discoideum" ''Bioinformatics'' 23:2543-49</ref>, Alveolata<ref>Zhu G, LaGier MJ, Stejskal F, Millership JJ, Cai X et al (2002) "Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase" ''Gene'' 298:79-89</ref>, and animals <ref>Castoe TA, Stephens T, Noonan BP, Calestani C (2007) "A novel group of type I polyketide synthases (PKS) in animals and the complex phylogenomics of PKSs" ''Gene'' 392:47-58</ref><ref>Calestani C, Rast JP, Davidson EH (2003) "Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening" ''Development'' 130:4587-96</ref>. | ||
| − | + | | | |
| − | + | ポリケチド合成酵素は、バクテリア、菌類、植物、変形菌、アルベオラータ(せん毛虫や渦鞭毛虫を含む原生動物)、動物に見出されています。 | |
| − | + | }} | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
{| class="wikitable" | {| class="wikitable" | ||
| − | !Species || Actinomycetes || Cyanobacteria || γ-Proteobacteria || Fungi || Alveolata | + | |+ {{Bilingual|分布のまとめ|Distribution Summary}} |
| + | ! Species || Actinomycetes || Cyanobacteria || γ-Proteobacteria || Fungi || Alveolata | ||
|- align="center" | |- align="center" | ||
!Type-I PKS | !Type-I PKS | ||
| Line 288: | Line 278: | ||
|} | |} | ||
| − | + | ;References | |
| − | + | <references/> | |
| − | + | ==={{Bilingual|タイプ|Type}}=== | |
| − | + | {{Twocolumn | |
| − | + | |There are three types of PKSs known to date. | |
| − | + | |ポリケチド合成酵素には3つのタイプがあります。}} | |
| − | + | ||
| − | + | ||
| − | === | + | ===={{Bilingual|タイプ I|Type I}}==== |
| − | + | {{Twocolumn| | |
| + | Multiple domains per protein (e.g. Erythromycin biosynthesis <ref> (2001) ''Nat Prod Rep'' 18:380</ref>) | ||
| + | * Bacterial type I is '''modular''', i.e., each domain (or module) catalyses a specific transformation. | ||
| + | * Fungal type I is '''iterative''', i.e., it reuses same active sites through multiple catalytic steps. | ||
| + | ** Non-reducing (NR) type produces aromatic polyketides | ||
| + | ** partially reducing type produces others. | ||
| + | | | ||
| + | 複数ドメインを持つタンパク質 (例. エリスロマイシン合成酵素)。 少なくとも 60 kb 以上の大きなクラスターを形成し大きな環状化合物を作る。構造の修飾はやや限定的。 | ||
| + | * バクテリアのタイプ I は、モジュール型 | ||
| + | * 菌類のタイプ I は反復型 | ||
| + | ** 非還元型 (NR) は芳香族ポリケチドを作る | ||
| + | ** 部分還元型 はそれ以外の構造を作る | ||
| + | }} | ||
| + | |||
| + | ===={{Bilingual|タイプ II|Type II}}==== | ||
| + | {{Twocolumn| | ||
| + | single domain per protein | ||
| + | * Three proteins (KSα, KSβ, ACP) are repeatedly used for carbon chain elongation, and the chain length is determined by another protein, CLF. | ||
| + | * In bacteria, products are aromatic (e.g. chiorotetracycline, pradimicin). | ||
| + | | | ||
| + | ドメインを一つしか持たないタンパク質 (例. ) 30 kb 程度の中程度クラスターをなし、芳香族化合物が多い。 構造の修飾は多様。 | ||
| + | * 三種のタンパク質 (KSα, KSβ, ACP) が炭素鎖の伸張に利用され、鎖の長さは CLF タンパク質が決定する | ||
| + | * バクテリアでは芳香環をもつ化合物を生産する (例. クロルテトラサイクリン、プラディマイシン) | ||
| + | }} | ||
| + | |||
| + | ===={{Bilingual|タイプ III|Type III}}==== | ||
| + | {{Twocolumn| | ||
| + | Chalcone synthase-like in plants | ||
| + | * Discovered in plants, but later found in bacteria<ref>Moore BS, Hopke JN (2001) Discovery of a new bacterial polyketide biosynthetic pathway ''Chembiochem'' 2:35-8</ref> | ||
| + | | | ||
| + | カルコン合成酵素型 (植物) | ||
| + | * もともと植物で発見されたがバクテリアも持つことが後に判明 | ||
| + | }} | ||
| + | |||
| + | ===={{Bilingual|例外的なPKS|Unusual Type}}==== | ||
| + | {{Twocolumn| | ||
| + | ;Bacterial but iterative type I PKS for aromatic polyketide | ||
| + | | | ||
| + | ;バクテリア由来の反復型タイプ I なのに芳香族ポリケチドを合成するもの | ||
| + | }} | ||
| + | * AviM for orsellinic acid biosynthesis (''Streptomyces viridochromogens'' Tu57)<ref>Gaisser S, Trefzer A, Stockert S, Kirschning A, Bechthold A (1997) Cloning of an avilamycin biosynthetic gene cluster from Streptomyces viridochromogenes Tu57. ''J Bacteriol'' 179:6271-8</ref> | ||
| + | * CalO5 for calicheamicin biosyntehsis (''Micromonospora echinospora'' ssp. ''calichenisis'')<ref>Whitwam RE, Ahlert J, Holman TR, Ruppen M, Thorson JS (2000) The gene calC encodes for a non-heme iron metalloprotein responsible for calicheamicin self-resistance in Micromonospora. ''J Am Chem Soc'' 122:1556-7</ref> | ||
| + | * NesB for neocarzinostatin biosynthesis (''?'')<ref>Zazopoulos E, Huang K, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM (2003) A genomics-guided approach for discovering and expressing cryptic metabolicpathways ''Nat Biotechnol'' epub.</ref> | ||
| + | |||
| + | {{Twocolumn| | ||
| + | ;Type I PKS that lacks the cognate AT domain | ||
| + | | | ||
| + | ;タイプ I PKS なのに AT ドメインが無いもの | ||
| + | }} | ||
| + | * lnmIJ for leinamycin biosynthesis (''Streptomyces atroolivaceus'' S-140)<ref>Cheng Y-Q, Tang G-L, Shen B (2003) Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis ''Proc Natl Acad Sci U S A'' 100: in press</ref> | ||
| + | * PedF for pederin biosynthesis (symbiont bacterium of Paederus beetles)<ref>Piel J (2002) A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles ''Proc Natl Acad Sci U S A'' 98:14808-13</ref> | ||
| + | |||
| + | {{Twocolumn| | ||
| + | ;Type II PKS that act non-iteratively and use acyl CoA as substrates directly | ||
| + | | | ||
| + | ;タイプ II PKS なのに反復型ではなく、アシルCoAを直接利用するもの | ||
| + | }} | ||
| + | * NonPQU and NonJK (''Streptomyces griseus'')<ref>Kwon HJ, Smith WC, Scharon AJ, Hwang SH, Kurth MJ, Shen B (2002) C-O bond formation by polyketide synthases ''Science'' 297(5585):1327-30</ref> | ||
| − | |||
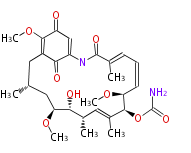
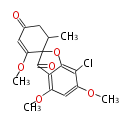
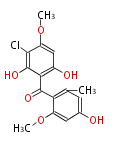
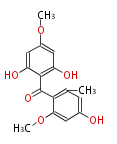
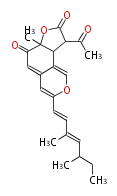
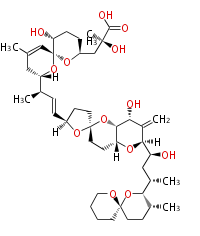
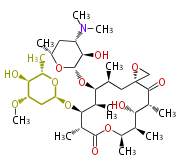
===Unusual structures=== | ===Unusual structures=== | ||
| Line 314: | Line 358: | ||
|} | |} | ||
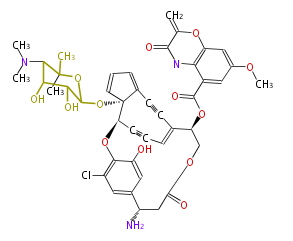
| − | == | + | ==Non-ribosomal peptide synthase (NRPS)== |
| − | + | Coupling with PKS and NRPS | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | * vancomycin () | |
| − | * | + | * leinamycin (Curr opin chem biol 7:285, 2003) |
| − | * | + | * pseurotin (chem bio chem 8:1736-1743, 2007) |
| + | * curacin (curr opin chem biol 13:216, 2009) | ||
| + | * epothilone | ||
| + | * rapamycin | ||
| − | + | ===Decoration=== | |
| − | + | deoxysugars | |
| + | |||
| + | deoxygenation, c-methylation, amination, n-methylation, ketosugar, | ||
| + | ; References | ||
<references/> | <references/> | ||
Revision as of 16:07, 4 June 2011
| Polyketide Top | Species List | UniRef90 Class | UniRef50 Class | Gene Class | Domains (by CDD) |
Domains (by MAPSI) |
Polyketide
|
Overview
Acetate-Malonate Pathway
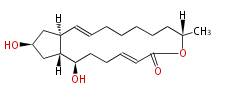
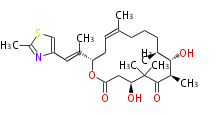
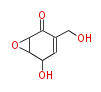
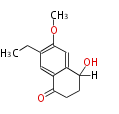
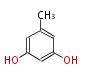
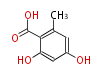
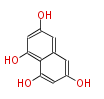
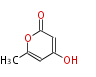
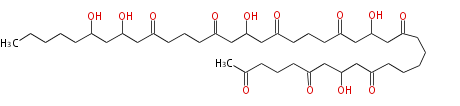
Acetyl CoA from glycolysis and its carboxylated form (malonyl CoA) are polymerized to form polyketone as in the fatty acid synthesis without reduction of carbonyl groups. Methylene moieties between ketones are highly reactive and easily Aldol- or Claisen-condensed to generate aromatic rings.
Elongation Mechanism
Polyketides are synthesized through the polymerization of acetyl units (β-ketomethylene) as in fatty acid biosynthesis. Typical starter units are short-chain fatty acids (e.g. acetyl-CoA or propionyl-CoA), on to which extender units (e.g. malonyl-CoA or methylmalonyl-CoA) are repeatedly polymerized. The key reactions for the chain extension are:
- Claisen condensation by β-ketoacyl synthase (KS)
- an acyltransferase (AT), and
- an acyl carrier protein (ACP).
After elongation, β-ketone is reduced. In fatty acid biosynthesis, the chain is fully reduced by the following three steps:
- Reduction to an alcohol by ketoreductase (KR),
- Dehydration to the conjugated ester by dehydratase (DH), and
- Reduction of the double bond by enoyl reductase (ER).
Finally, the chain is terminated by a thioesterase (TE) activity and allows Claisen cyclization (CYC).
Classification
3-4th digits
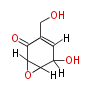
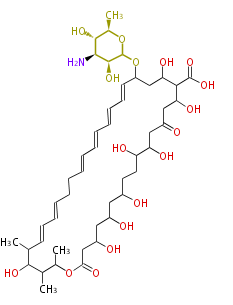
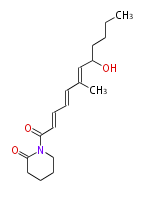
| Linear Chain and Related (L) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
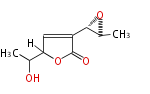
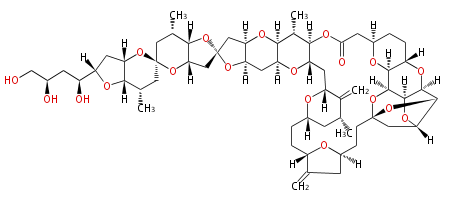
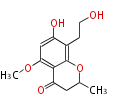
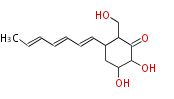
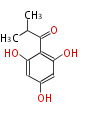
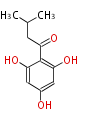
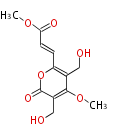
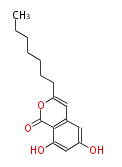
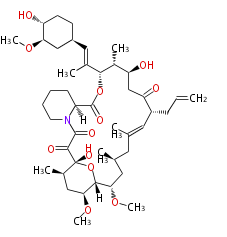
Acetogenins (LA) | ||||||||||||
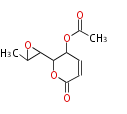
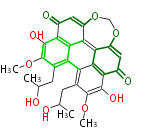
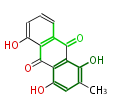
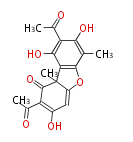
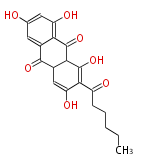
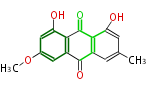
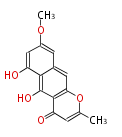
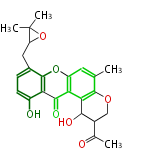
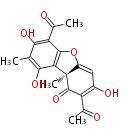
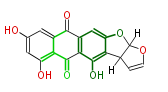
| Aromatic and Diels-Alder Related (most often by iterative type II) | ||||||||||||||
|
|
| ||||||||||||
|
| |||||||||||||
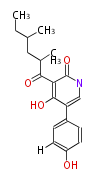
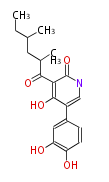
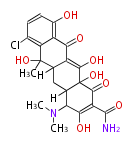
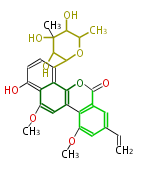
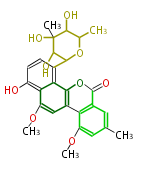
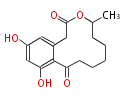
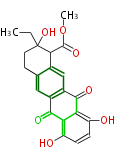
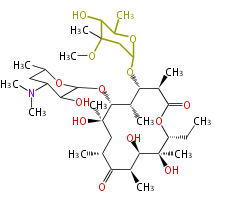
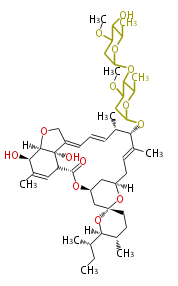
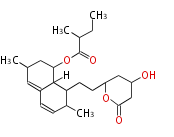
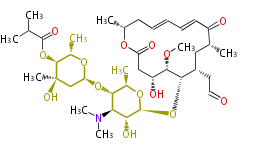
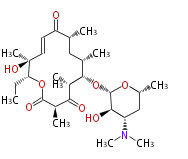
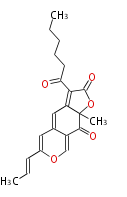
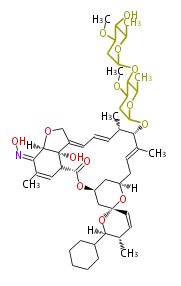
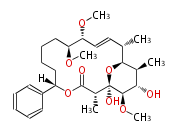
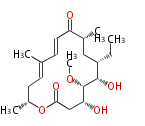
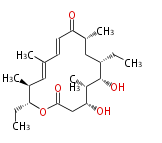
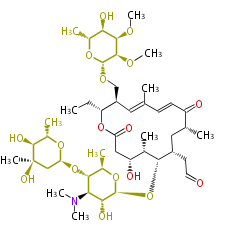
| Macrolides (most often by non-iterative type I) | ||||||||||||||
|
|
| ||||||||||||
|
|
| ||||||||||||
|
| |||||||||||||
- References
- ↑ foo
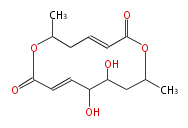
- ↑ 6-deoxy sugars (L-cladinose and D-desosamine) are attached.
- ↑ http://www.indiana.edu/~drwchem/pdfs/50.pdf
- ↑ =Pimaricin
5th digit
| The number of C2 unit | ||
|---|---|---|
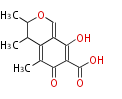
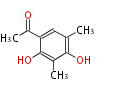
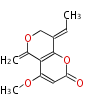
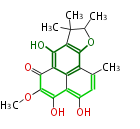
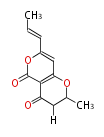
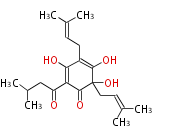
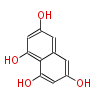
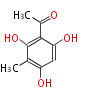
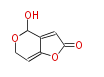
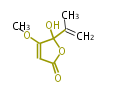
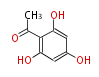
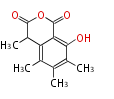
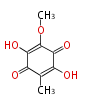
| 4 Units orsellinic acid, 6-methylsalicylic acid, triacetic acid lactone, asperlin, usnic acid, methylphloracetophenone, penicillic acid, patulin |
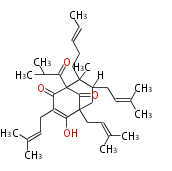
5 Units citrinin, aflatoxin, augenone, sepedonin, stipitatonic acid |
6 Units plumbagin, 7-methyljuglone, juglone, variotin |
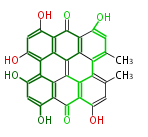
| 7 & 8 Units Anthraquinone rings |
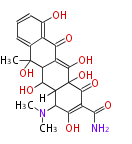
9 Units Tetracyclines |
>9 Units |
Polyketide Synthase (PKS)
Distribution
PKS members are found in bacteria, fungi, plants, slime mold[1], Alveolata[2], and animals [3][4].
| Species | Actinomycetes | Cyanobacteria | γ-Proteobacteria | Fungi | Alveolata |
|---|---|---|---|---|---|
| Type-I PKS | Ο | Ο | Ο | Ο | Ο |
| Type-II PKS | Ο | Χ | Χ | Χ | Χ |
| NRPS | Ο | Ο | Ο | Ο | Χ |
| deoxysugar | Ο | Χ | Χ | Χ | Χ |
| Terpene | Δ | Χ | Χ | Ο | Χ |
- References
- ↑ Zucko J, Skunca N, Curk T, Zupan B, Long PF et al (2007) "Polyketide synthase genes and the natural products potential of Dictyostelium discoideum" Bioinformatics 23:2543-49
- ↑ Zhu G, LaGier MJ, Stejskal F, Millership JJ, Cai X et al (2002) "Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase" Gene 298:79-89
- ↑ Castoe TA, Stephens T, Noonan BP, Calestani C (2007) "A novel group of type I polyketide synthases (PKS) in animals and the complex phylogenomics of PKSs" Gene 392:47-58
- ↑ Calestani C, Rast JP, Davidson EH (2003) "Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening" Development 130:4587-96
Type
There are three types of PKSs known to date.
Type I
Multiple domains per protein (e.g. Erythromycin biosynthesis [1])
- Bacterial type I is modular, i.e., each domain (or module) catalyses a specific transformation.
- Fungal type I is iterative, i.e., it reuses same active sites through multiple catalytic steps.
- Non-reducing (NR) type produces aromatic polyketides
- partially reducing type produces others.
Type II
single domain per protein
- Three proteins (KSα, KSβ, ACP) are repeatedly used for carbon chain elongation, and the chain length is determined by another protein, CLF.
- In bacteria, products are aromatic (e.g. chiorotetracycline, pradimicin).
Type III
Chalcone synthase-like in plants
- Discovered in plants, but later found in bacteria[2]
Unusual Type
- Bacterial but iterative type I PKS for aromatic polyketide
- AviM for orsellinic acid biosynthesis (Streptomyces viridochromogens Tu57)[3]
- CalO5 for calicheamicin biosyntehsis (Micromonospora echinospora ssp. calichenisis)[4]
- NesB for neocarzinostatin biosynthesis (?)[5]
- Type I PKS that lacks the cognate AT domain
- lnmIJ for leinamycin biosynthesis (Streptomyces atroolivaceus S-140)[6]
- PedF for pederin biosynthesis (symbiont bacterium of Paederus beetles)[7]
- Type II PKS that act non-iteratively and use acyl CoA as substrates directly
- NonPQU and NonJK (Streptomyces griseus)[8]
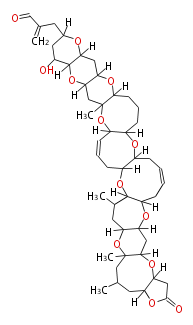
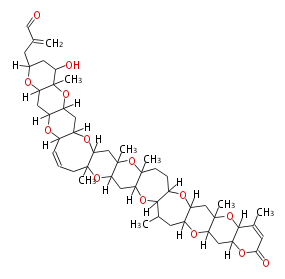
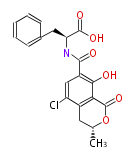
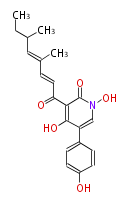
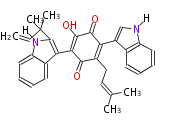
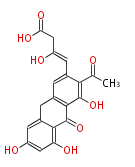
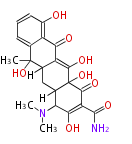
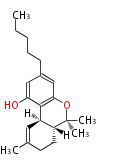
Unusual structures
| Phoma | zaragozic acid, phomoidoride | Streptomyces | yatakemycin, leinamycin, saframycin, neocarzinostatin, staurosporin, FR182877 | Other bacteria | PKS-NRPS hybrid type
Curacin A (Lyngbya), Shiphonazole (Herpetosiphon), Jamaicamide A (Lyngbya), Cylindrospermopsin (Cylindrospermopsis) |
|---|
Non-ribosomal peptide synthase (NRPS)
Coupling with PKS and NRPS
- vancomycin ()
- leinamycin (Curr opin chem biol 7:285, 2003)
- pseurotin (chem bio chem 8:1736-1743, 2007)
- curacin (curr opin chem biol 13:216, 2009)
- epothilone
- rapamycin
Decoration
deoxysugars
deoxygenation, c-methylation, amination, n-methylation, ketosugar,
- References
- ↑ (2001) Nat Prod Rep 18:380
- ↑ Moore BS, Hopke JN (2001) Discovery of a new bacterial polyketide biosynthetic pathway Chembiochem 2:35-8
- ↑ Gaisser S, Trefzer A, Stockert S, Kirschning A, Bechthold A (1997) Cloning of an avilamycin biosynthetic gene cluster from Streptomyces viridochromogenes Tu57. J Bacteriol 179:6271-8
- ↑ Whitwam RE, Ahlert J, Holman TR, Ruppen M, Thorson JS (2000) The gene calC encodes for a non-heme iron metalloprotein responsible for calicheamicin self-resistance in Micromonospora. J Am Chem Soc 122:1556-7
- ↑ Zazopoulos E, Huang K, Staffa A, Liu W, Bachmann BO, Nonaka K, Ahlert J, Thorson JS, Shen B, Farnet CM (2003) A genomics-guided approach for discovering and expressing cryptic metabolicpathways Nat Biotechnol epub.
- ↑ Cheng Y-Q, Tang G-L, Shen B (2003) Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis Proc Natl Acad Sci U S A 100: in press
- ↑ Piel J (2002) A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles Proc Natl Acad Sci U S A 98:14808-13
- ↑ Kwon HJ, Smith WC, Scharon AJ, Hwang SH, Kurth MJ, Shen B (2002) C-O bond formation by polyketide synthases Science 297(5585):1327-30