Category:PK
m (→Type I, II, and III) |
m |
||
| Line 1: | Line 1: | ||
{{PKS/Header}} | {{PKS/Header}} | ||
| − | {{Huge| | + | {{Huge|{{Bilingual|ポリケチド|Polyketide}}}} |
{| style="float:right" | {| style="float:right" | ||
| __TOC__ | | __TOC__ | ||
Revision as of 21:09, 19 December 2010
| Polyketide Top | Species List | UniRef90 Class | UniRef50 Class | Gene Class | Domains (by CDD) |
Domains (by MAPSI) |
Polyketide
|
Class Overview
Polyketides are synthesized through the polymerization of acetyl units (β-ketomethylene) as in fatty acid biosynthesis. Typical starter units are short-chain fatty acids (e.g. acetyl-CoA or propionyl-CoA), on to which extender units (e.g. malonyl-CoA or methylmalonyl-CoA) are repeatedly polymerized. The key reactions for the chain extension are:
- Claisen condensation by β-ketoacyl synthase (KS)
- an acyltransferase (AT), and
- an acyl carrier protein (ACP).
After elongation, β-ketone is reduced. In fatty acid biosynthesis, the chain is fully reduced by the following three steps:
- Reduction to an alcohol by ketoreductase (KR),
- Dehydration to the conjugated ester by dehydratase (DH), and
- Reduction of the double bond by enoyl reductase (ER).
Finally, the chain is terminated by a thioesterase (TE) activity and allows Claisen cyclization (CYC).
Classification
3-4th digits
| Linear Chain and Related (L) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
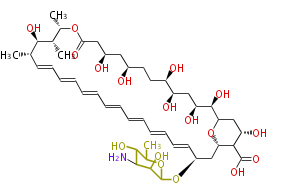
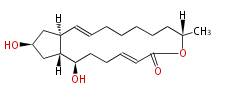
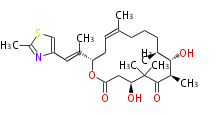
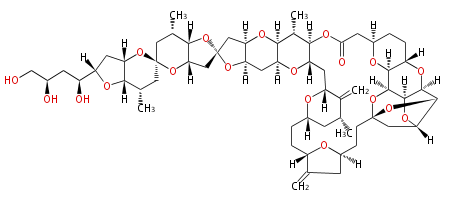
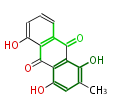
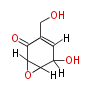
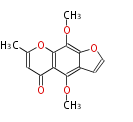
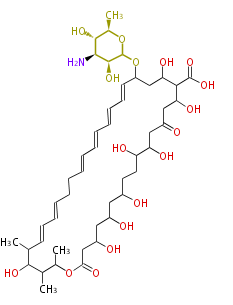
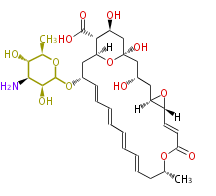
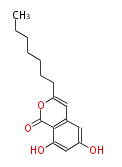
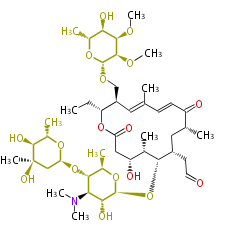
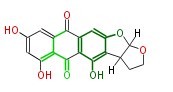
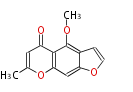
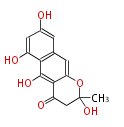
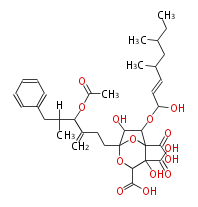
Acetogenins (LA) | ||||||||||||
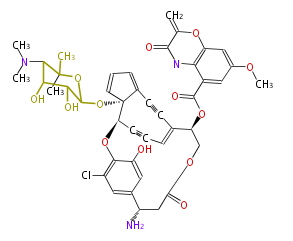
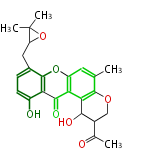
| Aromatic and Diels-Alder Related (most often by iterative type II) | ||||||||||||||
|
|
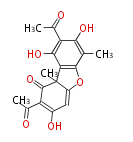
| ||||||||||||
|
| |||||||||||||
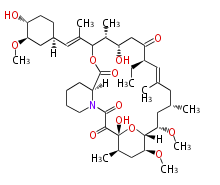
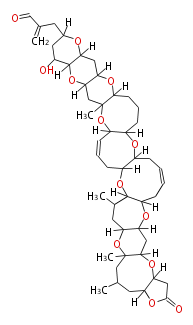
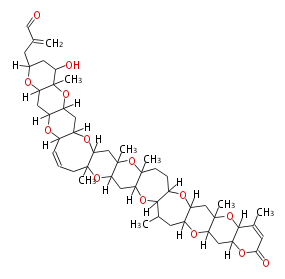
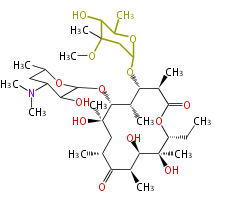
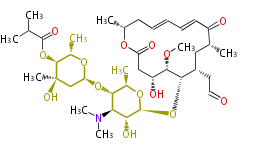
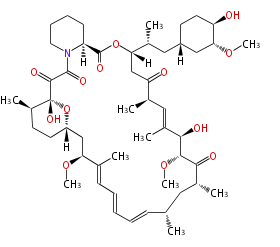
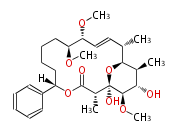
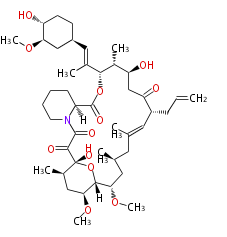
| Macrolides (most often by non-iterative type I) | ||||||||||||||
|
|
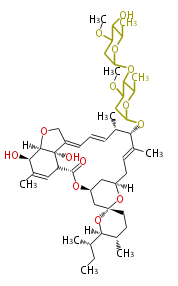
| ||||||||||||
|
|
| ||||||||||||
|
| |||||||||||||
- References
- ↑ foo
- ↑ 6-deoxy sugars (L-cladinose and D-desosamine) are attached.
- ↑ http://www.indiana.edu/~drwchem/pdfs/50.pdf
- ↑ =Pimaricin
5th digit
| The number of C2 unit | ||
|---|---|---|
| 4 Units orsellinic acid, 6-methylsalicylic acid, triacetic acid lactone, asperlin, usnic acid, methylphloracetophenone, penicillic acid, patulin |
5 Units citrinin, aflatoxin, augenone, sepedonin, stipitatonic acid |
6 Units plumbagin, 7-methyljuglone, juglone, variotin |
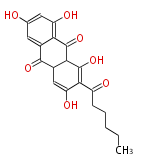
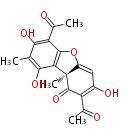
| 7 & 8 Units Anthraquinone rings |
9 Units Tetracyclines |
>9 Units |
Polyketide Synthase (PKS)
Distribution
PKS members are found in bacteria, fungi, plants, slime mold[1], Alveolata[2], and animals [3][4].
- References
- ↑ Zucko J, Skunca N, Curk T, Zupan B, Long PF et al (2007) "Polyketide synthase genes and the natural products potential of Dictyostelium discoideum" Bioinformatics 23:2543-49
- ↑ Zhu G, LaGier MJ, Stejskal F, Millership JJ, Cai X et al (2002) "Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase" Gene 298:79-89
- ↑ Castoe TA, Stephens T, Noonan BP, Calestani C (2007) "A novel group of type I polyketide synthases (PKS) in animals and the complex phylogenomics of PKSs" Gene 392:47-58
- ↑ Calestani C, Rast JP, Davidson EH (2003) "Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening" Development 130:4587-96
Type I, II, and III
There are three types of PKSs known to date.
Type I : multiple domains per protein (e.g. Erythromycin biosynthesis [1])
- Bacterial type I is modular.
- Fungal type I is "iterative", i.e., it reuses same active sites through multiple catalytic steps. Non-reducing (NR) type produces aromatic polyketides, and partially reducing type produces others.
Type II : single domain per protein
- Three proteins (KSα, KSβ, ACP) are repeatedly used for carbon chain elongation, and the chain length is determined by another protein, CLF.
- In bacteria, products are aromatic (e.g. chiorotetracycline, pradimicin).
Type III : chalcone synthase-like in plants
- Discovered in plants, but later found in bacteria[2]
| Species | Actinomycetes | Cyanobacteria | γ-Proteobacteria | Fungi | Alveolata |
|---|---|---|---|---|---|
| Type-I PKS | Ο | Ο | Ο | Ο | Ο |
| Type-II PKS | Ο | Χ | Χ | Χ | Χ |
| NRPS | Ο | Ο | Ο | Ο | Χ |
| deoxysugar | Ο | Χ | Χ | Χ | Χ |
| Terpene | Δ | Χ | Χ | Ο | Χ |
Non-ribosomal peptide synthase (NRPS)
Coupling with PKS and NRPS
- vancomycin ()
- leinamycin (Curr opin chem biol 7:285, 2003)
- pseurotin (chem bio chem 8:1736-1743, 2007)
- curacin (curr opin chem biol 13:216, 2009)
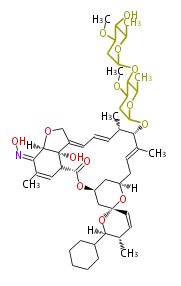
- epothilone
- rapamycin
Decoration
deoxysugars
deoxygenation, c-methylation, amination, n-methylation, ketosugar,
Unusual structures
| Phoma | zaragozic acid, phomoidoride | Streptomyces | yatakemycin, leinamycin, saframycin, neocarzinostatin, staurosporin, FR182877 | Other bacteria | PKS-NRPS hybrid type
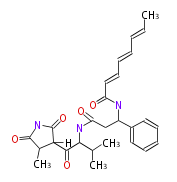
Curacin A (Lyngbya), Shiphonazole (Herpetosiphon), Jamaicamide A (Lyngbya), Cylindrospermopsin (Cylindrospermopsis) |
|---|
Cite error:
<ref> tags exist, but no <references/> tag was found