Category:FLI
(→{{Bilingual|耳にする名前|Familiar Names}}) |
|||
| Line 36: | Line 36: | ||
<references/> | <references/> | ||
| − | ==={{Bilingual| | + | ==={{Bilingual|代表例|Representative Names}}=== |
| − | + | {| style="vertical-align:top" | |
| − | + | | [[FLIA1ANS0002|daidzein]]<br/>[[Image:FLIA1ANS0002.png|120px]] | |
| − | + | | [[FLIAAANS0001|genistein]]<br/>[[Image:FLIAAANS0001.png|120px]] | |
| − | + | | [[FLIE1ANS0001|coumestrol]]<br/>[[Image:FLIE1ANS0001.png|120px]] | |
| − | </ | + | | [[FLID1ANS0001|Medicarpin]]<ref>The major phytoalexin in alfalfa (''Medicago sativa'')</ref><br/>[[Image:FLID1ANS0001.png|120px]] |
| + | |} | ||
| + | |||
| + | <references/> | ||
=={{Bilingual|生合成|Biosynthesis}}== | =={{Bilingual|生合成|Biosynthesis}}== | ||
Revision as of 12:07, 19 August 2010
Isoflavonoid
| Flavonoid Top | Molecule Index | Author Index | Journals | Structure Search | Food | New Input |
Upper classes : FL Flavonoid
Contents |
Overview
| 2nd Class | |||||||
|---|---|---|---|---|---|---|---|
| FLIA | Isoflavone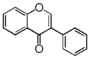
|
FLIB | Isoflavanone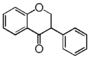
|
FLIC | Isoflavan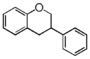
|
FLID | Pterocarpane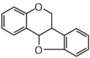
|
| FLIE | Coumestan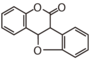
|
FLIF | Rotenoid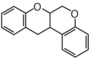
|
FLIG | Coumaranochromone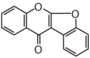
|
FLIH | 3-Arylcoumarin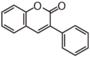
|
| FLII | 2-Arylbenzofuran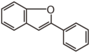
|
FLIJ | alpha-Methoxynbenzoin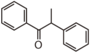
| ||||
The discovery of isoflavonoid was in 1842, when Reinsch and Hlasiwetz obtained ononin from the roots of spiny restharrow (Ononis spinosa L.)[1]. Its structure was later determined by Baker et al. as Formononetin 7-O-glucoside (1933). Since then, most isoflavonoids have been found in Fabaceae (bean family).
First isoflavonoid found in non-bean family was iridin from the rhizomes of Iris florentina (Iridaceae)[2]. The first structural identification was Prunetin from Prunus species in 1910 by Finnemore. Major isoflavonoids in iris are isoflavones and their O-glycosides. In iris, isoflavonoids are produced only in the rhizomes except for a trace of coumaranochromones in leaves [3].
- ↑ Reinsch H, Repert. Pharm. 26:12-31, 1842; Reinsch H, Repert. Pharm. 28:18-25, 1842; Hlasiwetz H, J. Prakt. Chem. 65:419-450, 1855; Hlasiwetz H, Sitzungsber. Kais. Akad. Wiss. Wien, Math.-Naturwiss. Kl. 15:142-168, 1855
- ↑ de Laire G and Tiemann F, Chem. Ber., 26:2010, 1893
- ↑ Iwashita T, Ootani S: Flavonoids of the genus Iris: structures, distribution and function (review). Ann. Tsukuba Bot. Gard. 17:147-183 (in Japanese)
Representative Names
daidzein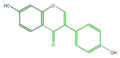
|
genistein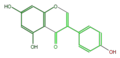
|
coumestrol
|
Medicarpin[1]
|
- ↑ The major phytoalexin in alfalfa (Medicago sativa)
Biosynthesis
This pathway is said to exist as an enzyme complex at ER membrane. [1]
| liquiritigenin |
IFS |
daidzein |
IOMT |
formononetin |
I2'H + IFR |
vestitone |
VR + DMID |
medicarpin |
| Structural Genes (continued from Flavonoid) | |||
|---|---|---|---|
| Abbrev. | Name | Origin | Information |
| IFS | isoflavone synthase | cytochrome P450 oxygenase family [2] | Use both liquiritigenin and naringenin as substrates to produce genistein and daidzein, respectively. |
| IOMT | isoflavone O-methyltransferase | ||
| I2'H | isoflavone 2'-hydroxylase | cytochrome P450 oxygenase family[5] | |
| IFR | isoflavone reductase | ||
| VR | vestitone reductase | ||
| DMID | 7,2'-dihydroxy,4'-methyxyisoflavanol dehydratase | ||
- ↑ Hrazdina G: Compartmentation in aromatic metabolism. In HA Stafford, RK Ibrahim, eds, Recent Advances in Phytochemistry. Plenum Press, New York, 1992 pp 1–23
- ↑ Akashi T, Aoki T, Ayabe S: Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol 1999 121:821-828
- ↑ Steele CL, Gijzen M, Qutob D, Dixon RA: Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch Biochem Biophys 1999 367: 146–150
- ↑ Jung W, Yu O, Sze-Mei CL, O’Keefe DP, Odell J, Fader G, McGonigle B: Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol 2000 18: 208–213
- ↑ Akashi T, Aoki T, Ayabe S: CYP81E1, a cytochrome P450 cDNA of licorice (Glycyrrhiza echinata L.), encodes isoflavone 29-hydroxylase. 1998 Biochem Biophys Res Comm 251: 67–70
Database statistics
Major Plant Families
|
|
The number in each family is counted as the number of genera (not species) listed in our registered references. Each reference record is accessible by clicking the link in compound pages. The taxonomy follows the APG-II classification. For details (or if the figure is broken), visit this page. 各科のカウントは種名でなく文献に記載された属名の数です。文献は代謝物ページのリンクからたどれ、分類はAPG-IIです。左の図が表示されない場合はここをクリックしてください。 |