Mol:PR100465
From Metabolomics.JP
ACD/Labs08070914192D
66 72 0 0 1 0 0 0 0 0 2 V2000
12.0627 -9.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.0627 -7.6207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.9427 -6.9207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.6827 -7.6207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.6827 -9.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.9427 -9.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
13.1827 -9.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.4427 -9.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.4427 -7.6207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
13.1827 -6.9207 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
10.9427 -11.1207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.4927 -6.9207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
15.6327 -6.9207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
16.8227 -7.6207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
18.0827 -6.9207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
18.0827 -5.5207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
16.8927 -4.8207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
15.6327 -5.5207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
19.2727 -4.8207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
15.6327 -9.7207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.7527 -11.8207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.5627 -11.1207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.3027 -11.8207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.3027 -13.2907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.4927 -13.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.7527 -13.2907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0427 -11.1207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8527 -11.8207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.0427 -13.9907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
8.4927 -15.3907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
10.9427 -13.9907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
19.2727 -7.6907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.8927 -3.4207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
20.5327 -6.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
16.8927 -10.4207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
16.8927 -11.7507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
18.0827 -12.4507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
19.2727 -11.7507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
19.2727 -10.4207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
18.0827 -9.7207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
20.5327 -12.4507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
15.6327 -12.5207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
20.5327 -9.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
18.0827 -13.8507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
21.7227 -10.3507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
22.9127 -9.7207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.1027 -10.3507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
25.2927 -9.6507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
25.2927 -8.2507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.1027 -7.6207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
22.9127 -8.3207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
24.1027 -6.2207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
26.4827 -7.5507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
26.4827 -10.4207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
24.1027 -11.7507 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
27.6890 -8.2952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
28.9337 -7.5683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
30.1384 -8.2427 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
31.3132 -7.5174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
27.7124 -9.3904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
32.5164 -8.1218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
33.7611 -7.3949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
33.7312 -5.9953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
32.5279 -5.3909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
31.2832 -6.1177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
34.9059 -5.2700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
1 6 1 0 0 0 0
1 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
2 10 1 0 0 0 0
6 11 1 0 0 0 0
4 12 1 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
13 18 1 0 0 0 0
16 19 1 0 0 0 0
8 20 1 0 0 0 0
21 11 1 1 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
21 26 1 0 0 0 0
23 27 1 1 0 0 0
27 28 1 0 0 0 0
24 29 1 6 0 0 0
25 30 1 1 0 0 0
26 31 1 6 0 0 0
15 32 1 0 0 0 0
17 33 1 0 0 0 0
32 34 1 0 0 0 0
35 20 1 6 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 40 1 0 0 0 0
35 40 1 0 0 0 0
38 41 1 1 0 0 0
36 42 1 1 0 0 0
39 43 1 6 0 0 0
37 44 1 6 0 0 0
43 45 1 0 0 0 0
46 45 1 6 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
49 50 1 0 0 0 0
50 51 1 0 0 0 0
46 51 1 0 0 0 0
50 52 1 1 0 0 0
49 53 1 6 0 0 0
48 54 1 1 0 0 0
47 55 1 1 0 0 0
53 56 1 0 0 0 0
56 57 1 0 0 0 0
57 58 2 0 0 0 0
58 59 1 0 0 0 0
56 60 2 0 0 0 0
59 61 2 0 0 0 0
61 62 1 0 0 0 0
62 63 2 0 0 0 0
63 64 1 0 0 0 0
64 65 2 0 0 0 0
59 65 1 0 0 0 0
63 66 1 0 0 0 0
M CHG 1 10 1
S SKP 7
CAS_RN 106863-71-2
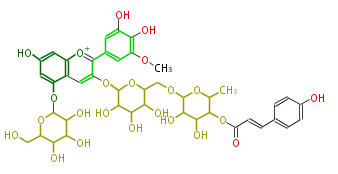
NAME Petunidin-3-O-(6''-O-(4'''-O-E-coum)-alpha-rhamnopyranosyl-beta-glucopyranosyl)-5-O-beta-glucopyranoside
ID PR100465
FORMULA C43H49O23
EXACTMASS 933.266462874
AVERAGEMASS 933.83536
SMILES OC(C(O)5)C(OC(COC(O7)C(O)C(C(C7C)OC(=O)C=Cc(c6)ccc(c6)O)O)C(O)5)Oc(c(c(c4)cc(c(O)c(OC)4)O)3)cc(c([o+1]3)1)c(OC(C(O)2)OC(CO)C(C2O)O)cc(c1)O
M END