Mol:LBF46000SC01
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 48 47 0 0 0 0 0 0 0 0999 V2000 | + | 48 47 0 0 0 0 0 0 0 0999 V2000 |
| − | 5.0009 0.8828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0009 0.8828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7172 1.2939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7172 1.2939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4322 0.8828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4322 0.8828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7172 2.1216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7172 2.1216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2872 1.2966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2872 1.2966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5709 0.8855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5709 0.8855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8559 1.3008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8559 1.3008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1395 0.8896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1395 0.8896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4272 1.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4272 1.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7109 0.8924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7109 0.8924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0014 1.3062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0014 1.3062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7178 0.8951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7178 0.8951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4300 1.3090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4300 1.3090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1464 0.8979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1464 0.8979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8586 1.3117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8586 1.3117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5750 0.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5750 0.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2872 1.3145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2872 1.3145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0036 0.9034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0036 0.9034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7159 1.3172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7159 1.3172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.4322 0.9061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.4322 0.9061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.4322 -0.1540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.4322 -0.1540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6760 -0.6215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6760 -0.6215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9610 -0.1540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9610 -0.1540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2584 -0.5885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2584 -0.5885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5681 -0.1375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5681 -0.1375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8655 -0.5720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8655 -0.5720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1739 -0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1739 -0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4712 -0.5541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4712 -0.5541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7824 -0.1031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7824 -0.1031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0797 -0.5376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0797 -0.5376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6119 -0.0853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6119 -0.0853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3145 -0.5197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3145 -0.5197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0034 -0.0688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0034 -0.0688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7060 -0.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7060 -0.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3976 -0.0509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3976 -0.0509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1003 -0.4854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1003 -0.4854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7891 -0.0344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7891 -0.0344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4917 -0.4689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4917 -0.4689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4917 -1.6115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4917 -1.6115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8290 -2.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8290 -2.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1415 -1.6115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1415 -1.6115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4664 -2.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4664 -2.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7830 -1.6225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7830 -1.6225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1079 -2.0983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1079 -2.0983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4272 -1.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4272 -1.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7521 -2.1106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7521 -2.1106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0688 -1.6459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0688 -1.6459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6064 -2.1216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6064 -2.1216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 2 4 2 0 0 0 0 | + | 2 4 2 0 0 0 0 |
| − | 1 5 1 0 0 0 0 | + | 1 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID LBF46000SC01 | + | ID LBF46000SC01 |
| − | FORMULA C46H92O2 | + | FORMULA C46H92O2 |
| − | EXACTMASS 676.709732188 | + | EXACTMASS 676.709732188 |
| − | AVERAGEMASS 677.22148 | + | AVERAGEMASS 677.22148 |
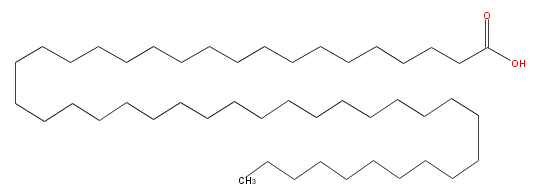
| − | SMILES C(CCCCCC(O)=O)CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | + | SMILES C(CCCCCC(O)=O)CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
48 47 0 0 0 0 0 0 0 0999 V2000
5.0009 0.8828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7172 1.2939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4322 0.8828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7172 2.1216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2872 1.2966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5709 0.8855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8559 1.3008 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1395 0.8896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4272 1.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7109 0.8924 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0014 1.3062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7178 0.8951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4300 1.3090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1464 0.8979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8586 1.3117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5750 0.9006 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2872 1.3145 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0036 0.9034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7159 1.3172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.4322 0.9061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.4322 -0.1540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6760 -0.6215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9610 -0.1540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2584 -0.5885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5681 -0.1375 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8655 -0.5720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1739 -0.1196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4712 -0.5541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7824 -0.1031 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0797 -0.5376 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6119 -0.0853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3145 -0.5197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0034 -0.0688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7060 -0.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3976 -0.0509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1003 -0.4854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7891 -0.0344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4917 -0.4689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4917 -1.6115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8290 -2.0790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1415 -1.6115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4664 -2.0872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7830 -1.6225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1079 -2.0983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4272 -1.6349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7521 -2.1106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0688 -1.6459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6064 -2.1216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
2 4 2 0 0 0 0
1 5 1 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 13 1 0 0 0 0
13 14 1 0 0 0 0
14 15 1 0 0 0 0
15 16 1 0 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 19 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
S SKP 5
ID LBF46000SC01
FORMULA C46H92O2
EXACTMASS 676.709732188
AVERAGEMASS 677.22148
SMILES C(CCCCCC(O)=O)CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC
M END