Mol:LBF38000SC01
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 40 39 0 0 0 0 0 0 0 0999 V2000 | + | 40 39 0 0 0 0 0 0 0 0999 V2000 |
| − | 4.7740 0.1327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7740 0.1327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4904 0.5438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4904 0.5438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.2054 0.1327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.2054 0.1327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4904 1.3716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4904 1.3716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0604 0.5466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0604 0.5466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3440 0.1354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3440 0.1354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6290 0.5507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6290 0.5507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9126 0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9126 0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2004 0.5534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2004 0.5534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4840 0.1423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4840 0.1423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2283 0.5562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2283 0.5562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9446 0.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9446 0.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6569 0.5589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6569 0.5589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3733 0.1478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3733 0.1478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0855 0.5617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0855 0.5617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8019 0.1506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8019 0.1506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5141 0.5644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5141 0.5644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2305 0.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2305 0.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9428 0.5672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9428 0.5672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6591 0.1561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6591 0.1561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6591 -0.9041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6591 -0.9041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9029 -1.3716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9029 -1.3716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1879 -0.9041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1879 -0.9041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4853 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4853 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7950 -0.8876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7950 -0.8876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0924 -1.3221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0924 -1.3221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4008 -0.8697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4008 -0.8697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6981 -1.3042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6981 -1.3042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0092 -0.8532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0092 -0.8532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3066 -1.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3066 -1.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3850 -0.8353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3850 -0.8353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0876 -1.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0876 -1.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7765 -0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7765 -0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4791 -1.2533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4791 -1.2533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1708 -0.8009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1708 -0.8009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8734 -1.2354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8734 -1.2354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5622 -0.7844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5622 -0.7844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2649 -1.2189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2649 -1.2189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9565 -0.7666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9565 -0.7666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.6591 -1.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.6591 -1.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 2 4 2 0 0 0 0 | + | 2 4 2 0 0 0 0 |
| − | 1 5 1 0 0 0 0 | + | 1 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID LBF38000SC01 | + | ID LBF38000SC01 |
| − | FORMULA C38H76O2 | + | FORMULA C38H76O2 |
| − | EXACTMASS 564.584531676 | + | EXACTMASS 564.584531676 |
| − | AVERAGEMASS 565.00884 | + | AVERAGEMASS 565.00884 |
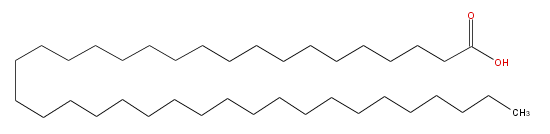
| − | SMILES CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC(O)=O | + | SMILES CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC(O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
40 39 0 0 0 0 0 0 0 0999 V2000
4.7740 0.1327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4904 0.5438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.2054 0.1327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4904 1.3716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0604 0.5466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3440 0.1354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6290 0.5507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9126 0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2004 0.5534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4840 0.1423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2283 0.5562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9446 0.1451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6569 0.5589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3733 0.1478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0855 0.5617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8019 0.1506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5141 0.5644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2305 0.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9428 0.5672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.6591 0.1561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.6591 -0.9041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9029 -1.3716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1879 -0.9041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4853 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7950 -0.8876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0924 -1.3221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4008 -0.8697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6981 -1.3042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0092 -0.8532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3066 -1.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3850 -0.8353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0876 -1.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7765 -0.8188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4791 -1.2533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1708 -0.8009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8734 -1.2354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5622 -0.7844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2649 -1.2189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9565 -0.7666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.6591 -1.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 1 0 0 0 0
2 4 2 0 0 0 0
1 5 1 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 13 1 0 0 0 0
13 14 1 0 0 0 0
14 15 1 0 0 0 0
15 16 1 0 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 19 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 40 1 0 0 0 0
S SKP 5
ID LBF38000SC01
FORMULA C38H76O2
EXACTMASS 564.584531676
AVERAGEMASS 565.00884
SMILES CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC(O)=O
M END