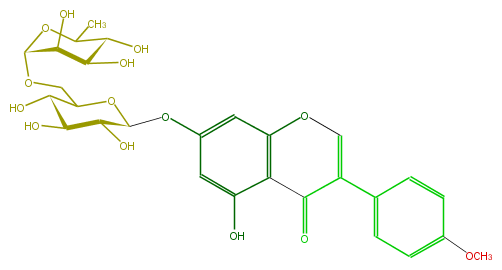
Mol:FLIAABGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.2013 0.4222 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2013 0.4222 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4805 0.0061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4805 0.0061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7597 0.4222 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7597 0.4222 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0391 0.0063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0391 0.0063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0391 -0.8567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0391 -0.8567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7082 -1.2882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7082 -1.2882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4555 -0.8567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4555 -0.8567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4555 0.0063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4555 0.0063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7082 0.4377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7082 0.4377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4802 -0.8257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4802 -0.8257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7597 -1.2418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7597 -1.2418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7082 -2.1505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7082 -2.1505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2023 -1.2878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2023 -1.2878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2023 -2.1128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2023 -2.1128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9168 -2.5252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9168 -2.5252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6311 -2.1128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6311 -2.1128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6311 -1.2878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6311 -1.2878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9168 -0.8754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9168 -0.8754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7733 -2.0720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7733 -2.0720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7299 2.2829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7299 2.2829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1674 1.7613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1674 1.7613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4901 1.8319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4901 1.8319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8739 1.5332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8739 1.5332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4361 2.0549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4361 2.0549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1135 1.9843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1135 1.9843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7663 2.3800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7663 2.3800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8011 1.8528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8011 1.8528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0915 1.5544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0915 1.5544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6517 0.8537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6517 0.8537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3010 0.1696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3010 0.1696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5977 0.3095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5977 0.3095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9614 0.2045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9614 0.2045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3414 0.7475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3414 0.7475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0563 0.6246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0563 0.6246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2496 0.6155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2496 0.6155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9344 0.2334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9344 0.2334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0819 -0.1865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0819 -0.1865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3893 1.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3893 1.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0852 1.1410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0852 1.1410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3432 2.5878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3432 2.5878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1850 -2.4945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1850 -2.4945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2496 -2.5878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2496 -2.5878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 5 1 0 0 0 0 | + | 11 5 1 0 0 0 0 |
| − | 6 12 2 0 0 0 0 | + | 6 12 2 0 0 0 0 |
| − | 7 13 1 0 0 0 0 | + | 7 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 11 19 1 0 0 0 0 | + | 11 19 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 24 27 1 0 0 0 0 | + | 24 27 1 0 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 21 39 1 0 0 0 0 | + | 21 39 1 0 0 0 0 |
| − | 39 38 1 0 0 0 0 | + | 39 38 1 0 0 0 0 |
| − | 22 40 1 0 0 0 0 | + | 22 40 1 0 0 0 0 |
| − | 32 1 1 0 0 0 0 | + | 32 1 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 16 41 1 0 0 0 0 | + | 16 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 46 -0.5539 0.3817 | + | M SBV 1 46 -0.5539 0.3817 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FLIAABGS0002 | + | ID FLIAABGS0002 |
| − | FORMULA C28H32O14 | + | FORMULA C28H32O14 |
| − | EXACTMASS 592.179205732 | + | EXACTMASS 592.179205732 |
| − | AVERAGEMASS 592.54528 | + | AVERAGEMASS 592.54528 |
| − | SMILES C(=C1c(c5)ccc(OC)c5)Oc(c2)c(c(cc2OC(O3)C(O)C(O)C(O)C(COC(O4)C(C(C(O)C(C)4)O)O)3)O)C1=O | + | SMILES C(=C1c(c5)ccc(OC)c5)Oc(c2)c(c(cc2OC(O3)C(O)C(O)C(O)C(COC(O4)C(C(C(O)C(C)4)O)O)3)O)C1=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-2.2013 0.4222 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4805 0.0061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7597 0.4222 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0391 0.0063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0391 -0.8567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7082 -1.2882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4555 -0.8567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4555 0.0063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7082 0.4377 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4802 -0.8257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7597 -1.2418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7082 -2.1505 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2023 -1.2878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2023 -2.1128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9168 -2.5252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6311 -2.1128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6311 -1.2878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9168 -0.8754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7733 -2.0720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7299 2.2829 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1674 1.7613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4901 1.8319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8739 1.5332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4361 2.0549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1135 1.9843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7663 2.3800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8011 1.8528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0915 1.5544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6517 0.8537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3010 0.1696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5977 0.3095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9614 0.2045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3414 0.7475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0563 0.6246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2496 0.6155 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9344 0.2334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0819 -0.1865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3893 1.0278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0852 1.1410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3432 2.5878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1850 -2.4945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2496 -2.5878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
2 10 1 0 0 0 0
10 11 2 0 0 0 0
11 5 1 0 0 0 0
6 12 2 0 0 0 0
7 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
11 19 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
25 26 1 0 0 0 0
24 27 1 0 0 0 0
23 28 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
34 38 1 0 0 0 0
21 39 1 0 0 0 0
39 38 1 0 0 0 0
22 40 1 0 0 0 0
32 1 1 0 0 0 0
41 42 1 0 0 0 0
16 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 OCH3
M SBV 1 46 -0.5539 0.3817
S SKP 5
ID FLIAABGS0002
FORMULA C28H32O14
EXACTMASS 592.179205732
AVERAGEMASS 592.54528
SMILES C(=C1c(c5)ccc(OC)c5)Oc(c2)c(c(cc2OC(O3)C(O)C(O)C(O)C(COC(O4)C(C(C(O)C(C)4)O)O)3)O)C1=O
M END