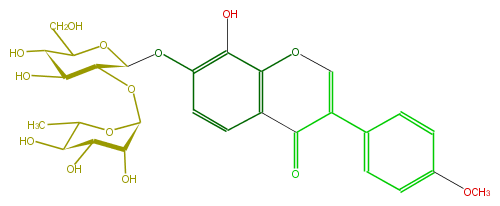
Mol:FLIA3AGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3746 1.0144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3746 1.0144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6537 0.5981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6537 0.5981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9328 1.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9328 1.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2122 0.5984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2122 0.5984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2122 -0.2647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2122 -0.2647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5353 -0.6962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5353 -0.6962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2826 -0.2647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2826 -0.2647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2826 0.5984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2826 0.5984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5353 1.0298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5353 1.0298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6534 -0.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6534 -0.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9328 -0.6498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9328 -0.6498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5353 -1.5586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5353 -1.5586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0294 -0.6959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0294 -0.6959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0294 -1.5209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0294 -1.5209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7440 -1.9333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7440 -1.9333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4584 -1.5209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4584 -1.5209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4584 -0.6959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4584 -0.6959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7440 -0.2833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7440 -0.2833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8122 1.0596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8122 1.0596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3634 0.4674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3634 0.4674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7173 0.7187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7173 0.7187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0440 0.7113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0440 0.7113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5469 1.1786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5469 1.1786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2070 0.9416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2070 0.9416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3123 1.0179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3123 1.0179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1879 0.5316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1879 0.5316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9540 0.2402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9540 0.2402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0787 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0787 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4160 -1.0579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4160 -1.0579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7674 -0.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7674 -0.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1148 -1.0579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1148 -1.0579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7774 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7774 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4261 -0.6589 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4261 -0.6589 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8556 -0.5251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8556 -0.5251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0990 -0.8552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0990 -0.8552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2531 -1.4500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2531 -1.4500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0900 -1.6725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0900 -1.6725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9328 1.8464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9328 1.8464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1901 -1.9433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1901 -1.9433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3123 -2.5912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3123 -2.5912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7199 1.5937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7199 1.5937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8927 2.5912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8927 2.5912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 5 1 0 0 0 0 | + | 11 5 1 0 0 0 0 |
| − | 6 12 2 0 0 0 0 | + | 6 12 2 0 0 0 0 |
| − | 7 13 1 0 0 0 0 | + | 7 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 22 1 1 0 0 0 0 | + | 22 1 1 0 0 0 0 |
| − | 3 38 1 0 0 0 0 | + | 3 38 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 16 39 1 0 0 0 0 | + | 16 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 24 41 1 0 0 0 0 | + | 24 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 44 -0.7317 0.4224 | + | M SBV 1 44 -0.7317 0.4224 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 46 0.5129 -0.6521 | + | M SBV 2 46 0.5129 -0.6521 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FLIA3AGS0001 | + | ID FLIA3AGS0001 |
| − | FORMULA C28H32O14 | + | FORMULA C28H32O14 |
| − | EXACTMASS 592.179205732 | + | EXACTMASS 592.179205732 |
| − | AVERAGEMASS 592.54528 | + | AVERAGEMASS 592.54528 |
| − | SMILES C(C(O)2)(CO)OC(Oc(c5O)ccc(c45)C(C(=CO4)c(c3)ccc(c3)OC)=O)C(C2O)OC(C1O)OC(C)C(C1O)O | + | SMILES C(C(O)2)(CO)OC(Oc(c5O)ccc(c45)C(C(=CO4)c(c3)ccc(c3)OC)=O)C(C2O)OC(C1O)OC(C)C(C1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-2.3746 1.0144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6537 0.5981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9328 1.0144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2122 0.5984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2122 -0.2647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5353 -0.6962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2826 -0.2647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2826 0.5984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5353 1.0298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6534 -0.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9328 -0.6498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5353 -1.5586 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0294 -0.6959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0294 -1.5209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7440 -1.9333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4584 -1.5209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4584 -0.6959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7440 -0.2833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8122 1.0596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3634 0.4674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7173 0.7187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0440 0.7113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5469 1.1786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2070 0.9416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3123 1.0179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1879 0.5316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9540 0.2402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0787 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4160 -1.0579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7674 -0.8725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1148 -1.0579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7774 -0.4736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4261 -0.6589 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8556 -0.5251 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0990 -0.8552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2531 -1.4500 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0900 -1.6725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9328 1.8464 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1901 -1.9433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3123 -2.5912 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7199 1.5937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8927 2.5912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
2 10 1 0 0 0 0
10 11 2 0 0 0 0
11 5 1 0 0 0 0
6 12 2 0 0 0 0
7 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
28 34 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 27 1 0 0 0 0
22 1 1 0 0 0 0
3 38 1 0 0 0 0
39 40 1 0 0 0 0
16 39 1 0 0 0 0
41 42 1 0 0 0 0
24 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 OCH3
M SBV 1 44 -0.7317 0.4224
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 ^ CH2OH
M SBV 2 46 0.5129 -0.6521
S SKP 5
ID FLIA3AGS0001
FORMULA C28H32O14
EXACTMASS 592.179205732
AVERAGEMASS 592.54528
SMILES C(C(O)2)(CO)OC(Oc(c5O)ccc(c45)C(C(=CO4)c(c3)ccc(c3)OC)=O)C(C2O)OC(C1O)OC(C)C(C1O)O
M END