Mol:FLIA1AGS0014
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 40 44 0 0 0 0 0 0 0 0999 V2000 | + | 40 44 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2121 1.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2121 1.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6558 1.4271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6558 1.4271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0995 1.7483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0995 1.7483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4566 1.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4566 1.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4566 0.7613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4566 0.7613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0333 0.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0333 0.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6101 0.7613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6101 0.7613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6101 1.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6101 1.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0333 1.7602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0333 1.7602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6556 0.7851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6556 0.7851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0995 0.4641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0995 0.4641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0333 -0.2372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0333 -0.2372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1864 0.4285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1864 0.4285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1864 -0.2081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1864 -0.2081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7378 -0.5264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7378 -0.5264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2891 -0.2081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2891 -0.2081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2891 0.4285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2891 0.4285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7378 0.7469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7378 0.7469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3573 1.3795 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.3573 1.3795 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.0110 0.9225 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.0110 0.9225 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5124 1.1164 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5124 1.1164 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9928 1.1107 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.9928 1.1107 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.3809 1.4713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3809 1.4713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8902 1.2884 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.8902 1.2884 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.1977 1.1543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1977 1.1543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5508 0.8962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5508 0.8962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2268 0.6368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2268 0.6368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5339 0.1048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5339 0.1048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0366 0.1048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0366 0.1048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5339 -0.4970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5339 -0.4970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0270 -0.7817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0270 -0.7817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0270 -1.3510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0270 -1.3510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5339 -1.6357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5339 -1.6357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0409 -1.3510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0409 -1.3510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0409 -0.7817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0409 -0.7817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5339 -2.2049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5339 -2.2049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4639 2.0321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4639 2.0321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8763 2.7466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8763 2.7466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1551 -0.7081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1551 -0.7081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0211 -1.2081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0211 -1.2081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 4 1 0 0 0 0 | + | 9 4 1 0 0 0 0 |
| − | 2 10 1 0 0 0 0 | + | 2 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 5 1 0 0 0 0 | + | 11 5 1 0 0 0 0 |
| − | 6 12 2 0 0 0 0 | + | 6 12 2 0 0 0 0 |
| − | 7 13 1 0 0 0 0 | + | 7 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 2 0 0 0 0 | + | 28 29 2 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 33 36 1 0 0 0 0 | + | 33 36 1 0 0 0 0 |
| − | 22 1 1 0 0 0 0 | + | 22 1 1 0 0 0 0 |
| − | 24 37 1 0 0 0 0 | + | 24 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 16 39 1 0 0 0 0 | + | 16 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 37 38 | + | M SAL 2 2 37 38 |
| − | M SBL 2 1 41 | + | M SBL 2 1 41 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 41 -3.0958 2.2049 | + | M SVB 2 41 -3.0958 2.2049 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 43 3.4832 -0.4144 | + | M SVB 1 43 3.4832 -0.4144 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FLIA1AGS0014 | + | ID FLIA1AGS0014 |
| − | KNApSAcK_ID C00010159 | + | KNApSAcK_ID C00010159 |
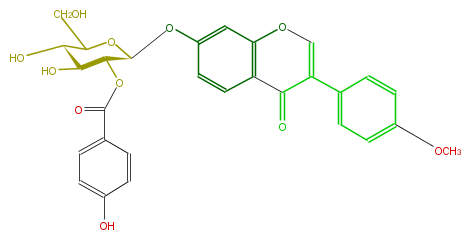
| − | NAME Formononetin 7-O-(2''-p-hydroxybenzoylglucoside) | + | NAME Formononetin 7-O-(2''-p-hydroxybenzoylglucoside) |
| − | CAS_RN 122130-27-2 | + | CAS_RN 122130-27-2 |
| − | FORMULA C29H26O11 | + | FORMULA C29H26O11 |
| − | EXACTMASS 550.147511674 | + | EXACTMASS 550.147511674 |
| − | AVERAGEMASS 550.51014 | + | AVERAGEMASS 550.51014 |
| − | SMILES OCC([C@H](O)1)O[C@@H](Oc(c3)ccc(C4=O)c(OC=C4c(c5)ccc(c5)OC)3)[C@@H](OC(c(c2)ccc(c2)O)=O)[C@H]1O | + | SMILES OCC([C@H](O)1)O[C@@H](Oc(c3)ccc(C4=O)c(OC=C4c(c5)ccc(c5)OC)3)[C@@H](OC(c(c2)ccc(c2)O)=O)[C@H]1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
40 44 0 0 0 0 0 0 0 0999 V2000
-1.2121 1.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6558 1.4271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0995 1.7483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4566 1.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4566 0.7613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0333 0.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6101 0.7613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6101 1.4272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0333 1.7602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6556 0.7851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0995 0.4641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0333 -0.2372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1864 0.4285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1864 -0.2081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7378 -0.5264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2891 -0.2081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2891 0.4285 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7378 0.7469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3573 1.3795 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.0110 0.9225 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5124 1.1164 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.9928 1.1107 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.3809 1.4713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8902 1.2884 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.1977 1.1543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5508 0.8962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2268 0.6368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5339 0.1048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0366 0.1048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5339 -0.4970 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0270 -0.7817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0270 -1.3510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5339 -1.6357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0409 -1.3510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0409 -0.7817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5339 -2.2049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4639 2.0321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8763 2.7466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1551 -0.7081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0211 -1.2081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 4 1 0 0 0 0
2 10 1 0 0 0 0
10 11 2 0 0 0 0
11 5 1 0 0 0 0
6 12 2 0 0 0 0
7 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
27 28 1 0 0 0 0
28 29 2 0 0 0 0
28 30 1 0 0 0 0
30 31 2 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 30 1 0 0 0 0
33 36 1 0 0 0 0
22 1 1 0 0 0 0
24 37 1 0 0 0 0
37 38 1 0 0 0 0
16 39 1 0 0 0 0
39 40 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 37 38
M SBL 2 1 41
M SMT 2 CH2OH
M SVB 2 41 -3.0958 2.2049
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 OCH3
M SVB 1 43 3.4832 -0.4144
S SKP 8
ID FLIA1AGS0014
KNApSAcK_ID C00010159
NAME Formononetin 7-O-(2''-p-hydroxybenzoylglucoside)
CAS_RN 122130-27-2
FORMULA C29H26O11
EXACTMASS 550.147511674
AVERAGEMASS 550.51014
SMILES OCC([C@H](O)1)O[C@@H](Oc(c3)ccc(C4=O)c(OC=C4c(c5)ccc(c5)OC)3)[C@@H](OC(c(c2)ccc(c2)O)=O)[C@H]1O
M END