Mol:FLIA1AGS0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 39 43 0 0 0 0 0 0 0 0999 V2000 | + | 39 43 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0254 0.0449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0254 0.0449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3109 0.4573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3109 0.4573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4033 0.0450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4033 0.0450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4033 -0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4033 -0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1442 -1.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1442 -1.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8849 -0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8849 -0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8849 0.0450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8849 0.0450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1442 0.4727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1442 0.4727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0254 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0254 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3109 -1.1921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3109 -1.1921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1442 -1.9524 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1442 -1.9524 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6251 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6251 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6251 -2.0555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6251 -2.0555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3333 -2.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3333 -2.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0414 -2.0555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0414 -2.0555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0414 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0414 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3333 -0.8289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3333 -0.8289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7223 -2.4486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7223 -2.4486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6819 0.4239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6819 0.4239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9878 0.2095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9878 0.2095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5277 -0.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5277 -0.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8652 -0.1401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8652 -0.1401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2260 -0.1332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2260 -0.1332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6905 0.3314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6905 0.3314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3673 0.0884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3673 0.0884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7223 0.0538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7223 0.0538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2551 -0.3420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2551 -0.3420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2368 -0.6549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2368 -0.6549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8646 0.6081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8646 0.6081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4934 2.0312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4934 2.0312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9149 1.5175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9149 1.5175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2765 1.5156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2765 1.5156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5918 2.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5918 2.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6065 2.4643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6065 2.4643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9149 2.0124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9149 2.0124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2765 1.9900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2765 1.9900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8646 1.3093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8646 1.3093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2765 0.9995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2765 0.9995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6161 1.7866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6161 1.7866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 7 2 0 0 0 0 | + | 6 7 2 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 3 1 0 0 0 0 | + | 8 3 1 0 0 0 0 |
| − | 1 9 1 0 0 0 0 | + | 1 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 2 0 0 0 0 | + | 5 11 2 0 0 0 0 |
| − | 6 12 1 0 0 0 0 | + | 6 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 15 1 0 0 0 0 | + | 18 15 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 19 23 1 0 0 0 0 | + | 19 23 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 30 1 0 0 0 0 | + | 34 30 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 30 37 1 0 0 0 0 | + | 30 37 1 0 0 0 0 |
| − | 37 29 1 0 0 0 0 | + | 37 29 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 43 0.0000 0.5160 | + | M SBV 1 43 0.0000 0.5160 |
| − | S SKP 5 | + | S SKP 5 |
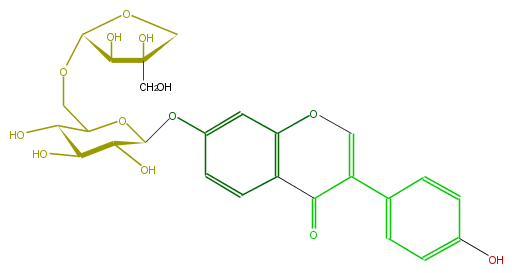
| − | ID FLIA1AGS0010 | + | ID FLIA1AGS0010 |
| − | FORMULA C26H28O13 | + | FORMULA C26H28O13 |
| − | EXACTMASS 548.152990982 | + | EXACTMASS 548.152990982 |
| − | AVERAGEMASS 548.49272 | + | AVERAGEMASS 548.49272 |
| − | SMILES C(C1OCC(O2)C(C(O)C(O)C2Oc(c3)cc(O5)c(C(=O)C(=C5)c(c4)ccc(O)c4)c3)O)(C(CO)(CO1)O)O | + | SMILES C(C1OCC(O2)C(C(O)C(O)C2Oc(c3)cc(O5)c(C(=O)C(=C5)c(c4)ccc(O)c4)c3)O)(C(CO)(CO1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
39 43 0 0 0 0 0 0 0 0999 V2000
-1.0254 0.0449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3109 0.4573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4033 0.0450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4033 -0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1442 -1.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8849 -0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8849 0.0450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1442 0.4727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0254 -0.7797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3109 -1.1921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1442 -1.9524 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6251 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6251 -2.0555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3333 -2.4643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0414 -2.0555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0414 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3333 -0.8289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7223 -2.4486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6819 0.4239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9878 0.2095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5277 -0.3978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8652 -0.1401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2260 -0.1332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6905 0.3314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3673 0.0884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7223 0.0538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2551 -0.3420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2368 -0.6549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8646 0.6081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4934 2.0312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9149 1.5175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2765 1.5156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5918 2.0308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6065 2.4643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9149 2.0124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2765 1.9900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8646 1.3093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2765 0.9995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6161 1.7866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 7 2 0 0 0 0
7 8 1 0 0 0 0
8 3 1 0 0 0 0
1 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 2 0 0 0 0
6 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 15 1 0 0 0 0
1 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
25 29 1 0 0 0 0
19 23 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 30 1 0 0 0 0
31 35 1 0 0 0 0
32 36 1 0 0 0 0
30 37 1 0 0 0 0
37 29 1 0 0 0 0
38 39 1 0 0 0 0
32 38 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 43
M SMT 1 CH2OH
M SBV 1 43 0.0000 0.5160
S SKP 5
ID FLIA1AGS0010
FORMULA C26H28O13
EXACTMASS 548.152990982
AVERAGEMASS 548.49272
SMILES C(C1OCC(O2)C(C(O)C(O)C2Oc(c3)cc(O5)c(C(=O)C(=C5)c(c4)ccc(O)c4)c3)O)(C(CO)(CO1)O)O
M END