Mol:FLIA1AGS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | 3.3121 -0.6042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3121 -0.6042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6584 -1.0612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6584 -1.0612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1570 -0.8673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1570 -0.8673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6766 -0.8731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6766 -0.8731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2885 -0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2885 -0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7792 -0.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7792 -0.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5437 -0.3768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5437 -0.3768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7848 -0.5125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7848 -0.5125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1141 -0.7559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1141 -0.7559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5865 1.1877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5865 1.1877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0302 0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0302 0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4738 1.1877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4738 1.1877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9178 0.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9178 0.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9178 0.2007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9178 0.2007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3410 -0.1323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3410 -0.1323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2357 0.2007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2357 0.2007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2357 0.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2357 0.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3410 1.1996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3410 1.1996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0299 0.2245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0299 0.2245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4738 -0.0966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4738 -0.0966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3410 -0.7978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3410 -0.7978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8121 -0.1321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8121 -0.1321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8121 -0.7687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8121 -0.7687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3634 -1.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3634 -1.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9148 -0.7687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9148 -0.7687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9148 -0.1321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9148 -0.1321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3634 0.1863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3634 0.1863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4661 -1.0870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4661 -1.0870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6003 1.0433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6003 1.0433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2291 0.5534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2291 0.5534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6946 0.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6946 0.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1788 0.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1788 0.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5536 1.1416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5536 1.1416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0212 0.8949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0212 0.8949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1141 0.7467 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1141 0.7467 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8077 0.5252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8077 0.5252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3883 0.2471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3883 0.2471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4197 1.6790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4197 1.6790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6228 1.4655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6228 1.4655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5207 -1.6790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5207 -1.6790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6957 -1.6790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6957 -1.6790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 1 0 0 0 | + | 1 2 1 1 0 0 0 |
| − | 2 3 1 1 0 0 0 | + | 2 3 1 1 0 0 0 |
| − | 4 3 1 1 0 0 0 | + | 4 3 1 1 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 5 8 1 0 0 0 0 | + | 5 8 1 0 0 0 0 |
| − | 4 9 1 0 0 0 0 | + | 4 9 1 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 11 19 1 0 0 0 0 | + | 11 19 1 0 0 0 0 |
| − | 19 20 2 0 0 0 0 | + | 19 20 2 0 0 0 0 |
| − | 20 14 1 0 0 0 0 | + | 20 14 1 0 0 0 0 |
| − | 15 21 2 0 0 0 0 | + | 15 21 2 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 2 0 0 0 0 | + | 24 25 2 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 2 0 0 0 0 | + | 26 27 2 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 25 28 1 0 0 0 0 | + | 25 28 1 0 0 0 0 |
| − | 1 28 1 0 0 0 0 | + | 1 28 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 10 32 1 0 0 0 0 | + | 10 32 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 3 40 1 0 0 0 0 | + | 3 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 42 | + | M SBL 1 1 42 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 42 -6.1971 5.6874 | + | M SBV 1 42 -6.1971 5.6874 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 44 | + | M SBL 2 1 44 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 44 -5.4350 4.0915 | + | M SBV 2 44 -5.4350 4.0915 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FLIA1AGS0005 | + | ID FLIA1AGS0005 |
| − | KNApSAcK_ID C00010080 | + | KNApSAcK_ID C00010080 |
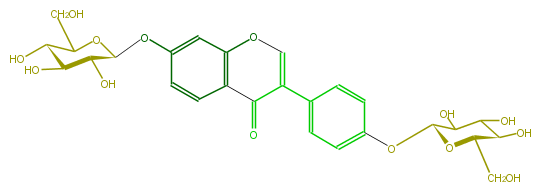
| − | NAME Daidzein 7,4'-di-O-glucoside | + | NAME Daidzein 7,4'-di-O-glucoside |
| − | CAS_RN 53681-67-7 | + | CAS_RN 53681-67-7 |
| − | FORMULA C27H30O14 | + | FORMULA C27H30O14 |
| − | EXACTMASS 578.163555668 | + | EXACTMASS 578.163555668 |
| − | AVERAGEMASS 578.5187000000001 | + | AVERAGEMASS 578.5187000000001 |
| − | SMILES O(c(c5)cc(c2c5)OC=C(c(c4)ccc(c4)OC(O3)C(O)C(O)C(O)C3CO)C2=O)C(C(O)1)OC(C(O)C(O)1)CO | + | SMILES O(c(c5)cc(c2c5)OC=C(c(c4)ccc(c4)OC(O3)C(O)C(O)C(O)C3CO)C2=O)C(C(O)1)OC(C(O)C(O)1)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
3.3121 -0.6042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6584 -1.0612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1570 -0.8673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6766 -0.8731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2885 -0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7792 -0.6954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5437 -0.3768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7848 -0.5125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1141 -0.7559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5865 1.1877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0302 0.8665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4738 1.1877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9178 0.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9178 0.2007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3410 -0.1323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2357 0.2007 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2357 0.8666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3410 1.1996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0299 0.2245 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4738 -0.0966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3410 -0.7978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8121 -0.1321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8121 -0.7687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3634 -1.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9148 -0.7687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9148 -0.1321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3634 0.1863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4661 -1.0870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6003 1.0433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2291 0.5534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6946 0.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1788 0.7668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5536 1.1416 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0212 0.8949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1141 0.7467 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8077 0.5252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3883 0.2471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4197 1.6790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6228 1.4655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5207 -1.6790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6957 -1.6790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 1 0 0 0
2 3 1 1 0 0 0
4 3 1 1 0 0 0
4 5 1 0 0 0 0
5 6 1 0 0 0 0
6 1 1 0 0 0 0
6 7 1 0 0 0 0
5 8 1 0 0 0 0
4 9 1 0 0 0 0
10 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
11 19 1 0 0 0 0
19 20 2 0 0 0 0
20 14 1 0 0 0 0
15 21 2 0 0 0 0
16 22 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
24 25 2 0 0 0 0
25 26 1 0 0 0 0
26 27 2 0 0 0 0
27 22 1 0 0 0 0
25 28 1 0 0 0 0
1 28 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
10 32 1 0 0 0 0
34 38 1 0 0 0 0
38 39 1 0 0 0 0
3 40 1 0 0 0 0
40 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 42
M SMT 1 ^CH2OH
M SBV 1 42 -6.1971 5.6874
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 44
M SMT 2 CH2OH
M SBV 2 44 -5.4350 4.0915
S SKP 8
ID FLIA1AGS0005
KNApSAcK_ID C00010080
NAME Daidzein 7,4'-di-O-glucoside
CAS_RN 53681-67-7
FORMULA C27H30O14
EXACTMASS 578.163555668
AVERAGEMASS 578.5187000000001
SMILES O(c(c5)cc(c2c5)OC=C(c(c4)ccc(c4)OC(O3)C(O)C(O)C(O)C3CO)C2=O)C(C(O)1)OC(C(O)C(O)1)CO
M END