Mol:FL7AAIGL0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.4628 1.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4628 1.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4628 0.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4628 0.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9065 0.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9065 0.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3502 0.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3502 0.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3502 1.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3502 1.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9065 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9065 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7939 0.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7939 0.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2376 0.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2376 0.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2376 1.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2376 1.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7939 1.4708 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -2.7939 1.4708 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6815 1.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6815 1.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1145 1.1434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1145 1.1434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5476 1.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5476 1.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5476 2.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5476 2.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1145 2.4528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1145 2.4528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6815 2.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6815 2.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0189 1.4707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0189 1.4707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3184 2.6254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3184 2.6254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9065 -0.4560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9065 -0.4560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8215 -0.2134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8215 -0.2134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3187 -0.2372 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.3187 -0.2372 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.5869 -0.7017 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.5869 -0.7017 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.0711 -0.5543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0711 -0.5543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4479 -0.7017 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.4479 -0.7017 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.7161 -0.2372 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.7161 -0.2372 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.2003 -0.3845 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.2003 -0.3845 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.6831 0.1274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6831 0.1274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3420 0.1442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3420 0.1442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9778 0.0747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9778 0.0747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8502 -1.1041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8502 -1.1041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6290 -1.9294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6290 -1.9294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0194 -2.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0194 -2.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6464 -2.2814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6464 -2.2814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3070 -2.7774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3070 -2.7774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0920 -3.3651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0920 -3.3651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2610 -3.9766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2610 -3.9766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0438 -4.5045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0438 -4.5045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2610 -5.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2610 -5.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8706 -5.0325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8706 -5.0325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1755 -4.5045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1755 -4.5045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8707 -3.9766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8707 -3.9766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1752 -5.5600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1752 -5.5600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8311 3.0289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8311 3.0289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3897 3.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3897 3.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4524 1.4707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4524 1.4707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4524 1.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4524 1.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | 15 43 1 0 0 0 0 | + | 15 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 13 45 1 0 0 0 0 | + | 13 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 45 46 | + | M SAL 2 2 45 46 |
| − | M SBL 2 1 49 | + | M SBL 2 1 49 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 49 -0.338 1.2645 | + | M SVB 2 49 -0.338 1.2645 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 47 -0.8311 3.0289 | + | M SVB 1 47 -0.8311 3.0289 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAIGL0012 | + | ID FL7AAIGL0012 |
| − | KNApSAcK_ID C00006903 | + | KNApSAcK_ID C00006903 |
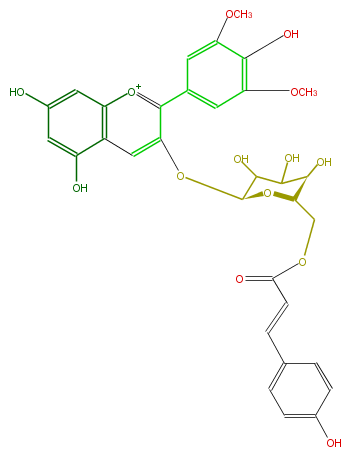
| − | NAME Malvidin 3-(6''-p-coumarylglucoside) | + | NAME Malvidin 3-(6''-p-coumarylglucoside) |
| − | CAS_RN 156577-22-9,158189-28-7 | + | CAS_RN 156577-22-9,158189-28-7 |
| − | FORMULA C32H31O14 | + | FORMULA C32H31O14 |
| − | EXACTMASS 639.1713807 | + | EXACTMASS 639.1713807 |
| − | AVERAGEMASS 639.58014 | + | AVERAGEMASS 639.58014 |
| − | SMILES O=C(OC[C@H]([C@H](O)2)O[C@@H](Oc(c4c(c5)cc(OC)c(c(OC)5)O)cc(c([o+1]4)3)c(O)cc(c3)O)C(O)C2O)C=Cc(c1)ccc(c1)O | + | SMILES O=C(OC[C@H]([C@H](O)2)O[C@@H](Oc(c4c(c5)cc(OC)c(c(OC)5)O)cc(c([o+1]4)3)c(O)cc(c3)O)C(O)C2O)C=Cc(c1)ccc(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-4.4628 1.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4628 0.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9065 0.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3502 0.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3502 1.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9065 1.4708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7939 0.1861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2376 0.5073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2376 1.1497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7939 1.4708 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
-1.6815 1.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1145 1.1434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5476 1.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5476 2.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1145 2.4528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6815 2.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0189 1.4707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3184 2.6254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9065 -0.4560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8215 -0.2134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3187 -0.2372 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.5869 -0.7017 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0711 -0.5543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4479 -0.7017 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.7161 -0.2372 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.2003 -0.3845 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.6831 0.1274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3420 0.1442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9778 0.0747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8502 -1.1041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6290 -1.9294 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0194 -2.2814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6464 -2.2814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3070 -2.7774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0920 -3.3651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2610 -3.9766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0438 -4.5045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2610 -5.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8706 -5.0325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1755 -4.5045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8707 -3.9766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1752 -5.5600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8311 3.0289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3897 3.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4524 1.4707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4524 1.4707 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
24 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
15 43 1 0 0 0 0
43 44 1 0 0 0 0
13 45 1 0 0 0 0
45 46 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 45 46
M SBL 2 1 49
M SMT 2 OCH3
M SVB 2 49 -0.338 1.2645
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 47
M SMT 1 OCH3
M SVB 1 47 -0.8311 3.0289
S SKP 8
ID FL7AAIGL0012
KNApSAcK_ID C00006903
NAME Malvidin 3-(6''-p-coumarylglucoside)
CAS_RN 156577-22-9,158189-28-7
FORMULA C32H31O14
EXACTMASS 639.1713807
AVERAGEMASS 639.58014
SMILES O=C(OC[C@H]([C@H](O)2)O[C@@H](Oc(c4c(c5)cc(OC)c(c(OC)5)O)cc(c([o+1]4)3)c(O)cc(c3)O)C(O)C2O)C=Cc(c1)ccc(c1)O
M END