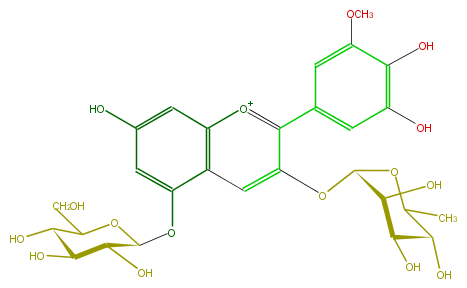
Mol:FL7AAHGO0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.5775 -0.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5775 -0.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5775 -1.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5775 -1.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8627 -1.5190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8627 -1.5190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1481 -1.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1481 -1.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1481 -0.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1481 -0.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8627 0.1316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8627 0.1316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5667 -1.5190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5667 -1.5190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2814 -1.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2814 -1.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2814 -0.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2814 -0.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5667 0.1316 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.5667 0.1316 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9958 0.1314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9958 0.1314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7242 -0.2892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7242 -0.2892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4527 0.1314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4527 0.1314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4527 0.9725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4527 0.9725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7242 1.3931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7242 1.3931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9958 0.9725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9958 0.9725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2919 0.1314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2919 0.1314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1577 1.3795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1577 1.3795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8627 -2.3440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8627 -2.3440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1622 -1.6182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1622 -1.6182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1056 -0.2457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1056 -0.2457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5569 -2.4561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5569 -2.4561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3512 -2.4561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3512 -2.4561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7803 -1.9038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7803 -1.9038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5851 -1.1293 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5851 -1.1293 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7908 -1.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7908 -1.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3616 -1.6815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3616 -1.6815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3111 -1.3996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3111 -1.3996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8874 -3.0286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8874 -3.0286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5171 -3.1891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5171 -3.1891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5452 -2.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5452 -2.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2636 -2.2741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2636 -2.2741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8187 -2.8614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8187 -2.8614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1782 -2.6123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1782 -2.6123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5105 -2.6196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5105 -2.6196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0092 -2.1564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0092 -2.1564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6636 -2.3914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6636 -2.3914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8832 -2.4630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8832 -2.4630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4835 -2.8185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4835 -2.8185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4590 -3.1473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4590 -3.1473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2849 -1.8056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2849 -1.8056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5452 -1.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5452 -1.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7242 2.0364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7242 2.0364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2914 3.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2914 3.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 13 21 1 0 0 0 0 | + | 13 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 19 1 0 0 0 0 | + | 35 19 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 15 43 1 0 0 0 0 | + | 15 43 1 0 0 0 0 |
| − | M CHG 1 10 1 | + | M CHG 1 10 1 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 46 0.6213 -0.5857 | + | M SBV 1 46 0.6213 -0.5857 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 48 0.0000 -0.6432 | + | M SBV 2 48 0.0000 -0.6432 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL7AAHGS0003 | + | ID FL7AAHGS0003 |
| − | FORMULA C28H33O16 | + | FORMULA C28H33O16 |
| − | EXACTMASS 625.176860008 | + | EXACTMASS 625.176860008 |
| − | AVERAGEMASS 625.55202 | + | AVERAGEMASS 625.55202 |
| − | SMILES OC(C1O)C(Oc(c52)cc(cc2[o+1]c(c(c5)OC(O4)C(O)C(O)C(C(C)4)O)c(c3)cc(c(O)c3O)OC)O)OC(C1O)CO | + | SMILES OC(C1O)C(Oc(c52)cc(cc2[o+1]c(c(c5)OC(O4)C(O)C(O)C(C(C)4)O)c(c3)cc(c(O)c3O)OC)O)OC(C1O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.5775 -0.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5775 -1.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8627 -1.5190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1481 -1.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1481 -0.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8627 0.1316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5667 -1.5190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2814 -1.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2814 -0.2811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5667 0.1316 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.9958 0.1314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7242 -0.2892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4527 0.1314 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4527 0.9725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7242 1.3931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9958 0.9725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2919 0.1314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1577 1.3795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8627 -2.3440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1622 -1.6182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1056 -0.2457 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5569 -2.4561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3512 -2.4561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7803 -1.9038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5851 -1.1293 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7908 -1.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3616 -1.6815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3111 -1.3996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8874 -3.0286 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5171 -3.1891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5452 -2.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2636 -2.2741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8187 -2.8614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1782 -2.6123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5105 -2.6196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0092 -2.1564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6636 -2.3914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8832 -2.4630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4835 -2.8185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4590 -3.1473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2849 -1.8056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5452 -1.5557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7242 2.0364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2914 3.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
13 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
20 26 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 19 1 0 0 0 0
41 42 1 0 0 0 0
37 41 1 0 0 0 0
43 44 1 0 0 0 0
15 43 1 0 0 0 0
M CHG 1 10 1
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 ^ CH2OH
M SBV 1 46 0.6213 -0.5857
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 48
M SMT 2 OCH3
M SBV 2 48 0.0000 -0.6432
S SKP 5
ID FL7AAHGS0003
FORMULA C28H33O16
EXACTMASS 625.176860008
AVERAGEMASS 625.55202
SMILES OC(C1O)C(Oc(c52)cc(cc2[o+1]c(c(c5)OC(O4)C(O)C(O)C(C(C)4)O)c(c3)cc(c(O)c3O)OC)O)OC(C1O)CO
M END