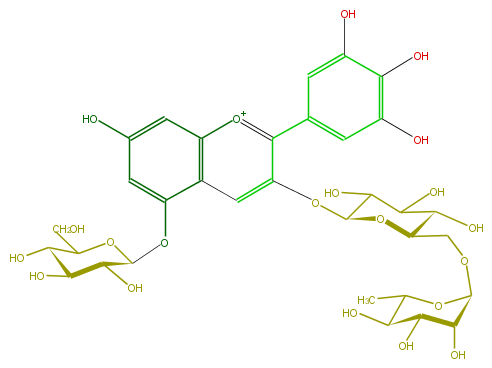
Mol:FL7AAGGL0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0211 0.8986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0211 0.8986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0211 0.0733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0211 0.0733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3064 -0.3394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3064 -0.3394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5916 0.0733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5916 0.0733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5916 0.8986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5916 0.8986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3064 1.3112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3064 1.3112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1232 -0.3394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1232 -0.3394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8379 0.0733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8379 0.0733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8379 0.8986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8379 0.8986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1232 1.3112 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.1232 1.3112 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5524 1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5524 1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2809 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2809 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0094 1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0094 1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0094 2.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0094 2.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2809 2.5729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2809 2.5729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5524 2.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5524 2.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2809 3.4139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2809 3.4139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7356 1.3111 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7356 1.3111 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7377 2.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7377 2.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3064 -1.1644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3064 -1.1644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7377 0.8907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7377 0.8907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7079 -0.3755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7079 -0.3755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6505 -1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6505 -1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2083 -1.8536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2083 -1.8536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5717 -1.6060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5717 -1.6060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9574 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9574 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4038 -1.1529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4038 -1.1529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0542 -1.3864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0542 -1.3864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1970 -1.4691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1970 -1.4691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7879 -1.8283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7879 -1.8283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9787 -2.0755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9787 -2.0755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4936 -2.2353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4936 -2.2353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1489 -2.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1489 -2.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8116 -2.6428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8116 -2.6428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4784 -2.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4784 -2.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8232 -2.2353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8232 -2.2353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1603 -2.4247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1603 -2.4247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8312 -2.2987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8312 -2.2987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8053 -0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8053 -0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3180 -0.7072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3180 -0.7072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0071 -0.6957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0071 -0.6957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6022 -1.0513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6022 -1.0513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0896 -0.5639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0896 -0.5639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4004 -0.5753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4004 -0.5753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1030 -0.1569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1030 -0.1569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0628 -0.1462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0628 -0.1462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8354 -0.8741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8354 -0.8741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3255 -1.0367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3255 -1.0367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6923 -1.5298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6923 -1.5298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4729 -2.5655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4729 -2.5655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4485 -3.2337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4485 -3.2337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4784 -3.4139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4784 -3.4139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5920 -0.8831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5920 -0.8831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8354 -0.5592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8354 -0.5592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 15 17 1 0 0 0 0 | + | 15 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 13 21 1 0 0 0 0 | + | 13 21 1 0 0 0 0 |
| − | 22 8 1 0 0 0 0 | + | 22 8 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 26 20 1 0 0 0 0 | + | 26 20 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 39 1 0 0 0 0 | + | 44 39 1 0 0 0 0 |
| − | 39 45 1 0 0 0 0 | + | 39 45 1 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 43 47 1 0 0 0 0 | + | 43 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 36 49 1 0 0 0 0 | + | 36 49 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 33 50 1 0 0 0 0 | + | 33 50 1 0 0 0 0 |
| − | 34 51 1 0 0 0 0 | + | 34 51 1 0 0 0 0 |
| − | 35 52 1 0 0 0 0 | + | 35 52 1 0 0 0 0 |
| − | 40 22 1 0 0 0 0 | + | 40 22 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 28 53 1 0 0 0 0 | + | 28 53 1 0 0 0 0 |
| − | M CHG 1 10 1 | + | M CHG 1 10 1 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 59 0.5379 -0.5033 | + | M SBV 1 59 0.5379 -0.5033 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL7AAGGL0012 | + | ID FL7AAGGL0012 |
| − | FORMULA C33H41O21 | + | FORMULA C33H41O21 |
| − | EXACTMASS 773.214033374 | + | EXACTMASS 773.214033374 |
| − | AVERAGEMASS 773.66604 | + | AVERAGEMASS 773.66604 |
| − | SMILES O(C(C6O)OC(C(C6O)O)COC(C5O)OC(C(O)C(O)5)C)c(c(c(c4)cc(O)c(O)c(O)4)3)cc(c2[o+1]3)c(cc(c2)O)OC(O1)C(O)C(C(O)C(CO)1)O | + | SMILES O(C(C6O)OC(C(C6O)O)COC(C5O)OC(C(O)C(O)5)C)c(c(c(c4)cc(O)c(O)c(O)4)3)cc(c2[o+1]3)c(cc(c2)O)OC(O1)C(O)C(C(O)C(CO)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-2.0211 0.8986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0211 0.0733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3064 -0.3394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5916 0.0733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5916 0.8986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3064 1.3112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1232 -0.3394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8379 0.0733 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8379 0.8986 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1232 1.3112 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.5524 1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2809 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0094 1.3111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0094 2.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2809 2.5729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5524 2.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2809 3.4139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7356 1.3111 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7377 2.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3064 -1.1644 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7377 0.8907 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7079 -0.3755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6505 -1.2700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2083 -1.8536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5717 -1.6060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9574 -1.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4038 -1.1529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0542 -1.3864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1970 -1.4691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7879 -1.8283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9787 -2.0755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4936 -2.2353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1489 -2.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8116 -2.6428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4784 -2.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8232 -2.2353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1603 -2.4247 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8312 -2.2987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8053 -0.2197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3180 -0.7072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0071 -0.6957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6022 -1.0513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0896 -0.5639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4004 -0.5753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1030 -0.1569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0628 -0.1462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8354 -0.8741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3255 -1.0367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6923 -1.5298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4729 -2.5655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4485 -3.2337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4784 -3.4139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5920 -0.8831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8354 -0.5592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
15 17 1 0 0 0 0
1 18 1 0 0 0 0
14 19 1 0 0 0 0
3 20 1 0 0 0 0
13 21 1 0 0 0 0
22 8 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 20 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 1 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 39 1 0 0 0 0
39 45 1 0 0 0 0
44 46 1 0 0 0 0
43 47 1 0 0 0 0
42 48 1 0 0 0 0
36 49 1 0 0 0 0
48 49 1 0 0 0 0
33 50 1 0 0 0 0
34 51 1 0 0 0 0
35 52 1 0 0 0 0
40 22 1 0 0 0 0
53 54 1 0 0 0 0
28 53 1 0 0 0 0
M CHG 1 10 1
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 59
M SMT 1 ^ CH2OH
M SBV 1 59 0.5379 -0.5033
S SKP 5
ID FL7AAGGL0012
FORMULA C33H41O21
EXACTMASS 773.214033374
AVERAGEMASS 773.66604
SMILES O(C(C6O)OC(C(C6O)O)COC(C5O)OC(C(O)C(O)5)C)c(c(c(c4)cc(O)c(O)c(O)4)3)cc(c2[o+1]3)c(cc(c2)O)OC(O1)C(O)C(C(O)C(CO)1)O
M END