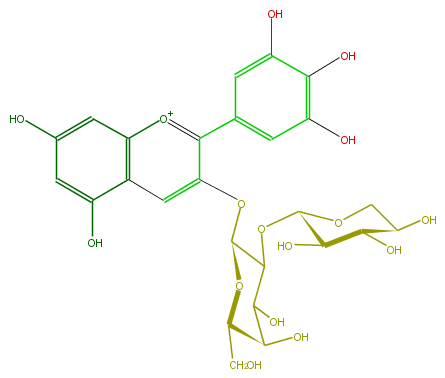
Mol:FL7AAGGA0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.3313 1.2642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3313 1.2642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3313 0.4393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3313 0.4393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6168 0.0268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6168 0.0268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9023 0.4393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9023 0.4393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9023 1.2642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9023 1.2642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6168 1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6168 1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1879 0.0268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1879 0.0268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4734 0.4393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4734 0.4393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4734 1.2642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4734 1.2642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1879 1.6767 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -1.1879 1.6767 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2408 1.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2408 1.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9690 1.2562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9690 1.2562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6971 1.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6971 1.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6971 2.5174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6971 2.5174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9690 2.9378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9690 2.9378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2408 2.5174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2408 2.5174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0455 1.6766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0455 1.6766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4251 2.9376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4251 2.9376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6168 -0.7979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6168 -0.7979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3717 -0.0133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3717 -0.0133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9690 3.7784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9690 3.7784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4251 1.2564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4251 1.2564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8199 -1.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8199 -1.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1687 -0.7894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1687 -0.7894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3371 -1.6640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3371 -1.6640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1128 -2.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1128 -2.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8199 -2.8622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8199 -2.8622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5955 -2.0770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5955 -2.0770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7650 -0.5299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7650 -0.5299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0125 -2.3810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0125 -2.3810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4977 -2.6727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4977 -2.6727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5078 -0.1954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5078 -0.1954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0237 -0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0237 -0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7666 -0.5875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7666 -0.5875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4835 -0.5798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4835 -0.5798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9625 -0.0587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9625 -0.0587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3126 -0.4018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3126 -0.4018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3238 -0.8550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3238 -0.8550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3587 -0.9322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3587 -0.9322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0455 -0.3325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0455 -0.3325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1949 -3.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1949 -3.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1124 -3.7784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1124 -3.7784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 13 22 1 0 0 0 0 | + | 13 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | 29 32 1 0 0 0 0 | + | 29 32 1 0 0 0 0 |
| − | 24 20 1 0 0 0 0 | + | 24 20 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 26 41 1 0 0 0 0 | + | 26 41 1 0 0 0 0 |
| − | M CHG 1 10 1 | + | M CHG 1 10 1 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 46 -0.0821 0.7948 | + | M SBV 1 46 -0.0821 0.7948 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL7AAGGA0002 | + | ID FL7AAGGA0002 |
| − | FORMULA C26H29O16 | + | FORMULA C26H29O16 |
| − | EXACTMASS 597.1455598800001 | + | EXACTMASS 597.1455598800001 |
| − | AVERAGEMASS 597.4988599999999 | + | AVERAGEMASS 597.4988599999999 |
| − | SMILES C(C(O)1)(C(OC(C5O)OCC(C5O)O)C(Oc(c2)c(c(c4)cc(O)c(c(O)4)O)[o+1]c(c3)c(c(cc3O)O)2)OC1CO)O | + | SMILES C(C(O)1)(C(OC(C5O)OCC(C5O)O)C(Oc(c2)c(c(c4)cc(O)c(c(O)4)O)[o+1]c(c3)c(c(cc3O)O)2)OC1CO)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-3.3313 1.2642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3313 0.4393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6168 0.0268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9023 0.4393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9023 1.2642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6168 1.6767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1879 0.0268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4734 0.4393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4734 1.2642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1879 1.6767 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.2408 1.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9690 1.2562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6971 1.6766 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6971 2.5174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9690 2.9378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2408 2.5174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0455 1.6766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4251 2.9376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6168 -0.7979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3717 -0.0133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9690 3.7784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4251 1.2564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8199 -1.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1687 -0.7894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3371 -1.6640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1128 -2.4538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8199 -2.8622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5955 -2.0770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7650 -0.5299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0125 -2.3810 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4977 -2.6727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5078 -0.1954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0237 -0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7666 -0.5875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4835 -0.5798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9625 -0.0587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3126 -0.4018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3238 -0.8550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3587 -0.9322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0455 -0.3325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1949 -3.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1124 -3.7784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
15 21 1 0 0 0 0
13 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
35 40 1 0 0 0 0
29 32 1 0 0 0 0
24 20 1 0 0 0 0
41 42 1 0 0 0 0
26 41 1 0 0 0 0
M CHG 1 10 1
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 CH2OH
M SBV 1 46 -0.0821 0.7948
S SKP 5
ID FL7AAGGA0002
FORMULA C26H29O16
EXACTMASS 597.1455598800001
AVERAGEMASS 597.4988599999999
SMILES C(C(O)1)(C(OC(C5O)OCC(C5O)O)C(Oc(c2)c(c(c4)cc(O)c(c(O)4)O)[o+1]c(c3)c(c(cc3O)O)2)OC1CO)O
M END