Mol:FL7AACGL0115
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 72 78 0 0 0 0 0 0 0 0999 V2000 | + | 72 78 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.7917 1.5969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7917 1.5969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7917 0.7719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7917 0.7719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5062 0.3594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5062 0.3594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2207 0.7719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2207 0.7719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2207 1.5969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2207 1.5969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5062 2.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5062 2.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9351 0.3594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9351 0.3594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6496 0.7719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6496 0.7719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6496 1.5969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6496 1.5969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9351 2.0094 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 2.9351 2.0094 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4261 2.0452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4261 2.0452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1406 1.6327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1406 1.6327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8551 2.0452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8551 2.0452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8551 2.8702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8551 2.8702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1406 3.2827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1406 3.2827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4261 2.8702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4261 2.8702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4953 3.2398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4953 3.2398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2938 0.4000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2938 0.4000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2117 1.9317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2117 1.9317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5062 -0.4033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5062 -0.4033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5000 1.6729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5000 1.6729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1268 -0.6448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1268 -0.6448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.9059 -0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.9059 -0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.8490 0.4509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.8490 0.4509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.2196 1.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.2196 1.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4405 0.9160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4405 0.9160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4973 0.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4973 0.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8925 0.0474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8925 0.0474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5116 -1.2095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5116 -1.2095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5598 -0.7370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5598 -0.7370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.4458 0.1632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.4458 0.1632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6708 -0.3275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6708 -0.3275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5163 1.9373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5163 1.9373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0695 1.2434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0695 1.2434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2823 1.4910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2823 1.4910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4787 1.3028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4787 1.3028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9254 1.9968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9254 1.9968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7128 1.7493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7128 1.7493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8979 2.3268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8979 2.3268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9755 1.4312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9755 1.4312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5048 1.4947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5048 1.4947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4225 0.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4225 0.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5003 2.6546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5003 2.6546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9122 3.3681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9122 3.3681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5003 4.0816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5003 4.0816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7361 3.3681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7361 3.3681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1481 2.6546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1481 2.6546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9719 2.6546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9719 2.6546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3844 3.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3844 3.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.2094 3.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.2094 3.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6219 2.6546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6219 2.6546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.2094 1.9401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.2094 1.9401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3844 1.9401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3844 1.9401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.4458 2.6546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.4458 2.6546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.6214 4.0826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.6214 4.0826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9581 -1.6548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9581 -1.6548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7734 -1.7821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7734 -1.7821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1117 -1.0294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1117 -1.0294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7863 -0.5540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7863 -0.5540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9709 -0.4266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9709 -0.4266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6326 -1.1794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6326 -1.1794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0780 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0780 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0310 -2.3343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0310 -2.3343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2902 -1.9689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2902 -1.9689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5023 -1.5645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5023 -1.5645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4949 -4.0826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4949 -4.0826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0837 -3.3687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0837 -3.3687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2598 -3.3679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2598 -3.3679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4963 -2.6555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4963 -2.6555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0850 -1.9417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0850 -1.9417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2611 -1.9409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2611 -1.9409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4976 -1.2285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4976 -1.2285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 8 18 1 0 0 0 0 | + | 8 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 13 21 1 0 0 0 0 | + | 13 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 32 28 1 0 0 0 0 | + | 32 28 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 35 42 1 0 0 0 0 | + | 35 42 1 0 0 0 0 |
| − | 19 36 1 0 0 0 0 | + | 19 36 1 0 0 0 0 |
| − | 25 21 1 0 0 0 0 | + | 25 21 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 53 48 1 0 0 0 0 | + | 53 48 1 0 0 0 0 |
| − | 51 54 1 0 0 0 0 | + | 51 54 1 0 0 0 0 |
| − | 50 55 1 0 0 0 0 | + | 50 55 1 0 0 0 0 |
| − | 43 39 1 0 0 0 0 | + | 43 39 1 0 0 0 0 |
| − | 56 57 1 1 0 0 0 | + | 56 57 1 1 0 0 0 |
| − | 57 58 1 1 0 0 0 | + | 57 58 1 1 0 0 0 |
| − | 59 58 1 1 0 0 0 | + | 59 58 1 1 0 0 0 |
| − | 59 60 1 0 0 0 0 | + | 59 60 1 0 0 0 0 |
| − | 60 61 1 0 0 0 0 | + | 60 61 1 0 0 0 0 |
| − | 61 56 1 0 0 0 0 | + | 61 56 1 0 0 0 0 |
| − | 61 62 1 0 0 0 0 | + | 61 62 1 0 0 0 0 |
| − | 56 63 1 0 0 0 0 | + | 56 63 1 0 0 0 0 |
| − | 57 64 1 0 0 0 0 | + | 57 64 1 0 0 0 0 |
| − | 58 65 1 0 0 0 0 | + | 58 65 1 0 0 0 0 |
| − | 59 18 1 0 0 0 0 | + | 59 18 1 0 0 0 0 |
| − | 66 67 2 0 0 0 0 | + | 66 67 2 0 0 0 0 |
| − | 67 68 1 0 0 0 0 | + | 67 68 1 0 0 0 0 |
| − | 67 69 1 0 0 0 0 | + | 67 69 1 0 0 0 0 |
| − | 69 70 1 0 0 0 0 | + | 69 70 1 0 0 0 0 |
| − | 70 71 2 0 0 0 0 | + | 70 71 2 0 0 0 0 |
| − | 70 72 1 0 0 0 0 | + | 70 72 1 0 0 0 0 |
| − | 72 62 1 0 0 0 0 | + | 72 62 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AACGL0115 | + | ID FL7AACGL0115 |
| − | KNApSAcK_ID C00014776 | + | KNApSAcK_ID C00014776 |
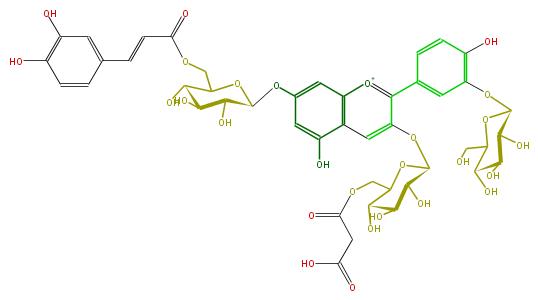
| − | NAME Cyanidin 3-(6-malonylglucoside)-7-(6-caffeoylglucoside)-3'-glucoside | + | NAME Cyanidin 3-(6-malonylglucoside)-7-(6-caffeoylglucoside)-3'-glucoside |
| − | CAS_RN 215176-96-8 | + | CAS_RN 215176-96-8 |
| − | FORMULA C45H49O27 | + | FORMULA C45H49O27 |
| − | EXACTMASS 1021.2461213619999 | + | EXACTMASS 1021.2461213619999 |
| − | AVERAGEMASS 1021.8543599999999 | + | AVERAGEMASS 1021.8543599999999 |
| − | SMILES Oc(c1O)cc(C=CC(OCC(O2)C(O)C(O)C(O)C2Oc(c7)cc(O)c(c57)cc(OC(O6)C(O)C(C(C6COC(=O)CC(O)=O)O)O)c([o+1]5)c(c3)ccc(c3OC(C(O)4)OC(C(C4O)O)CO)O)=O)cc1 | + | SMILES Oc(c1O)cc(C=CC(OCC(O2)C(O)C(O)C(O)C2Oc(c7)cc(O)c(c57)cc(OC(O6)C(O)C(C(C6COC(=O)CC(O)=O)O)O)c([o+1]5)c(c3)ccc(c3OC(C(O)4)OC(C(C4O)O)CO)O)=O)cc1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
72 78 0 0 0 0 0 0 0 0999 V2000
0.7917 1.5969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7917 0.7719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5062 0.3594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2207 0.7719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2207 1.5969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5062 2.0094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9351 0.3594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6496 0.7719 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6496 1.5969 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9351 2.0094 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
4.4261 2.0452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1406 1.6327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8551 2.0452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8551 2.8702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1406 3.2827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4261 2.8702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4953 3.2398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2938 0.4000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2117 1.9317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5062 -0.4033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.5000 1.6729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.1268 -0.6448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.9059 -0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.8490 0.4509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.2196 1.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.4405 0.9160 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.4973 0.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8925 0.0474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5116 -1.2095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.5598 -0.7370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.4458 0.1632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6708 -0.3275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5163 1.9373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0695 1.2434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2823 1.4910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4787 1.3028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9254 1.9968 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7128 1.7493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8979 2.3268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9755 1.4312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5048 1.4947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4225 0.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5003 2.6546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9122 3.3681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5003 4.0816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7361 3.3681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1481 2.6546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9719 2.6546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3844 3.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.2094 3.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.6219 2.6546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.2094 1.9401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3844 1.9401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.4458 2.6546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.6214 4.0826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9581 -1.6548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7734 -1.7821 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1117 -1.0294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7863 -0.5540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9709 -0.4266 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6326 -1.1794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0780 -0.9339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0310 -2.3343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2902 -1.9689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5023 -1.5645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4949 -4.0826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0837 -3.3687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2598 -3.3679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4963 -2.6555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0850 -1.9417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2611 -1.9409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4976 -1.2285 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
8 18 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
13 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
32 28 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
38 39 1 0 0 0 0
33 40 1 0 0 0 0
34 41 1 0 0 0 0
35 42 1 0 0 0 0
19 36 1 0 0 0 0
25 21 1 0 0 0 0
43 44 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 52 1 0 0 0 0
52 53 2 0 0 0 0
53 48 1 0 0 0 0
51 54 1 0 0 0 0
50 55 1 0 0 0 0
43 39 1 0 0 0 0
56 57 1 1 0 0 0
57 58 1 1 0 0 0
59 58 1 1 0 0 0
59 60 1 0 0 0 0
60 61 1 0 0 0 0
61 56 1 0 0 0 0
61 62 1 0 0 0 0
56 63 1 0 0 0 0
57 64 1 0 0 0 0
58 65 1 0 0 0 0
59 18 1 0 0 0 0
66 67 2 0 0 0 0
67 68 1 0 0 0 0
67 69 1 0 0 0 0
69 70 1 0 0 0 0
70 71 2 0 0 0 0
70 72 1 0 0 0 0
72 62 1 0 0 0 0
S SKP 8
ID FL7AACGL0115
KNApSAcK_ID C00014776
NAME Cyanidin 3-(6-malonylglucoside)-7-(6-caffeoylglucoside)-3'-glucoside
CAS_RN 215176-96-8
FORMULA C45H49O27
EXACTMASS 1021.2461213619999
AVERAGEMASS 1021.8543599999999
SMILES Oc(c1O)cc(C=CC(OCC(O2)C(O)C(O)C(O)C2Oc(c7)cc(O)c(c57)cc(OC(O6)C(O)C(C(C6COC(=O)CC(O)=O)O)O)c([o+1]5)c(c3)ccc(c3OC(C(O)4)OC(C(C4O)O)CO)O)=O)cc1
M END