Mol:FL7AACGL0107
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.4432 2.6741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4432 2.6741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4432 1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4432 1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7287 1.4366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7287 1.4366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0142 1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0142 1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0142 2.6741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0142 2.6741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7287 3.0866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7287 3.0866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2998 1.4366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2998 1.4366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4147 1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4147 1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4147 2.6741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4147 2.6741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2998 3.0866 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.2998 3.0866 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1912 3.1224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1912 3.1224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9057 2.7099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9057 2.7099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6202 3.1224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6202 3.1224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6202 3.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6202 3.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9057 4.3599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9057 4.3599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1912 3.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1912 3.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2604 4.3171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2604 4.3171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0589 1.4772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0589 1.4772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0231 3.0090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0231 3.0090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7287 0.8742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7287 0.8742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2802 2.7414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2802 2.7414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2898 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2898 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4886 -0.7653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4886 -0.7653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9587 -0.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9587 -0.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7088 0.2571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7088 0.2571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9304 0.5314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9304 0.5314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4602 -0.1469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4602 -0.1469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5194 -0.4710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5194 -0.4710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3490 -1.2593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3490 -1.2593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0899 -0.9562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0899 -0.9562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1229 0.2525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1229 0.2525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3737 -2.6992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3737 -2.6992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1969 -2.6414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1969 -2.6414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3587 -1.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3587 -1.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9103 -1.2183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9103 -1.2183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0870 -1.2760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0870 -1.2760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9252 -2.0853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9252 -2.0853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3298 -1.9697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3298 -1.9697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1295 -2.2797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1295 -2.2797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5964 -3.3453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5964 -3.3453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7675 -2.9313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7675 -2.9313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8588 -2.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8588 -2.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6302 0.0680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6302 0.0680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9272 -0.7735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9272 -0.7735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3990 -1.3018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3990 -1.3018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8347 -0.7570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8347 -0.7570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2802 -1.5985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2802 -1.5985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8070 -2.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8070 -2.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2178 -2.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2178 -2.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8036 -3.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8036 -3.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9786 -3.6474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9786 -3.6474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5678 -2.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5678 -2.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9820 -2.2185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9820 -2.2185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5650 -4.3599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5650 -4.3599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 8 18 1 0 0 0 0 | + | 8 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 13 21 1 0 0 0 0 | + | 13 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 25 18 1 0 0 0 0 | + | 25 18 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 32 40 1 0 0 0 0 | + | 32 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 35 28 1 0 0 0 0 | + | 35 28 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 53 48 1 0 0 0 0 | + | 53 48 1 0 0 0 0 |
| − | 51 54 1 0 0 0 0 | + | 51 54 1 0 0 0 0 |
| − | 43 31 1 0 0 0 0 | + | 43 31 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AACGL0107 | + | ID FL7AACGL0107 |
| − | KNApSAcK_ID C00014763 | + | KNApSAcK_ID C00014763 |
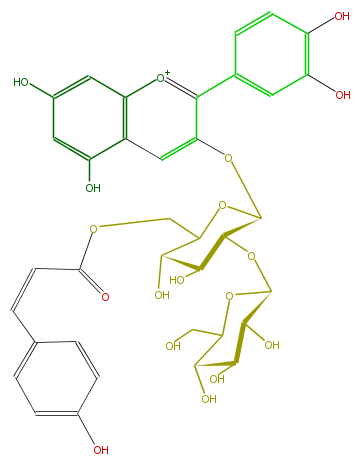
| − | NAME Cyanidin 3-(6''-(Z)-p-coumarylsophoroside) | + | NAME Cyanidin 3-(6''-(Z)-p-coumarylsophoroside) |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C36H37O18 | + | FORMULA C36H37O18 |
| − | EXACTMASS 757.1979893800001 | + | EXACTMASS 757.1979893800001 |
| − | AVERAGEMASS 757.66818 | + | AVERAGEMASS 757.66818 |
| − | SMILES C(C=Cc(c6)ccc(c6)O)(=O)OCC(O2)C(O)C(C(C2Oc(c4c(c5)cc(O)c(c5)O)cc(c3[o+1]4)c(cc(c3)O)O)OC(C(O)1)OC(CO)C(C(O)1)O)O | + | SMILES C(C=Cc(c6)ccc(c6)O)(=O)OCC(O2)C(O)C(C(C2Oc(c4c(c5)cc(O)c(c5)O)cc(c3[o+1]4)c(cc(c3)O)O)OC(C(O)1)OC(CO)C(C(O)1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-2.4432 2.6741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4432 1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7287 1.4366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0142 1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0142 2.6741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7287 3.0866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2998 1.4366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4147 1.8491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4147 2.6741 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2998 3.0866 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.1912 3.1224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9057 2.7099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6202 3.1224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6202 3.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9057 4.3599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1912 3.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2604 4.3171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0589 1.4772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0231 3.0090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7287 0.8742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2802 2.7414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2898 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4886 -0.7653 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9587 -0.0870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7088 0.2571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9304 0.5314 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4602 -0.1469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5194 -0.4710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3490 -1.2593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0899 -0.9562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1229 0.2525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3737 -2.6992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1969 -2.6414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3587 -1.8322 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9103 -1.2183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0870 -1.2760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9252 -2.0853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3298 -1.9697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1295 -2.2797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5964 -3.3453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7675 -2.9313 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8588 -2.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6302 0.0680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9272 -0.7735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3990 -1.3018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8347 -0.7570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2802 -1.5985 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8070 -2.2205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2178 -2.9359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8036 -3.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9786 -3.6474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5678 -2.9320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9820 -2.2185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5650 -4.3599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
8 18 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
13 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
24 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
27 31 1 0 0 0 0
25 18 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
32 40 1 0 0 0 0
33 41 1 0 0 0 0
34 42 1 0 0 0 0
35 28 1 0 0 0 0
43 44 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 52 1 0 0 0 0
52 53 2 0 0 0 0
53 48 1 0 0 0 0
51 54 1 0 0 0 0
43 31 1 0 0 0 0
S SKP 8
ID FL7AACGL0107
KNApSAcK_ID C00014763
NAME Cyanidin 3-(6''-(Z)-p-coumarylsophoroside)
CAS_RN -
FORMULA C36H37O18
EXACTMASS 757.1979893800001
AVERAGEMASS 757.66818
SMILES C(C=Cc(c6)ccc(c6)O)(=O)OCC(O2)C(O)C(C(C2Oc(c4c(c5)cc(O)c(c5)O)cc(c3[o+1]4)c(cc(c3)O)O)OC(C(O)1)OC(CO)C(C(O)1)O)O
M END