Mol:FL7AACGL0058
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.1465 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1465 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1465 -1.9810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1465 -1.9810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5902 -2.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5902 -2.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0339 -1.9810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0339 -1.9810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0339 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0339 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5902 -1.0174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5902 -1.0174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4775 -2.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4775 -2.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0788 -1.9810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0788 -1.9810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0788 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0788 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4775 -1.0174 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.4775 -1.0174 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6348 -1.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6348 -1.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2018 -1.3449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2018 -1.3449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7688 -1.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7688 -1.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7688 -0.3629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7688 -0.3629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2018 -0.0355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2018 -0.0355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6348 -0.3629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6348 -0.3629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7026 -1.0176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7026 -1.0176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3356 -0.0356 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3356 -0.0356 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5902 -2.9443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5902 -2.9443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4948 -2.7017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4948 -2.7017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2018 0.6190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2018 0.6190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7554 -2.3604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7554 -2.3604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4546 -2.8814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4546 -2.8814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0331 -2.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0331 -2.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6150 -2.8814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6150 -2.8814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9159 -2.3604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9159 -2.3604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3374 -2.5256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3374 -2.5256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1967 -2.5101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1967 -2.5101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5756 -2.1131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5756 -2.1131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6192 -2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6192 -2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9532 1.6798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9532 1.6798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5211 1.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5211 1.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3409 1.9824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3409 1.9824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5211 2.6168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5211 2.6168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9532 2.9447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9532 2.9447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1333 2.3141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1333 2.3141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3155 3.3070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3155 3.3070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5211 3.0260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5211 3.0260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4696 2.7039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4696 2.7039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7585 1.7413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7585 1.7413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1220 2.1738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1220 2.1738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1220 1.4250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1220 1.4250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3295 1.1643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3295 1.1643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8119 1.4250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8119 1.4250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1668 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1668 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7802 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7802 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1017 1.3672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1017 1.3672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7447 1.3672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7447 1.3672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0662 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0662 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7447 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7447 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1017 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1017 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7084 0.8104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7084 0.8104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0658 1.9233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0658 1.9233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9939 -2.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9939 -2.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7084 -3.3070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7084 -3.3070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 20 1 0 0 0 0 | + | 23 20 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 36 39 1 0 0 0 0 | + | 36 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 32 21 1 0 0 0 0 | + | 32 21 1 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 46 1 0 0 0 0 | + | 51 46 1 0 0 0 0 |
| − | 49 52 1 0 0 0 0 | + | 49 52 1 0 0 0 0 |
| − | 48 53 1 0 0 0 0 | + | 48 53 1 0 0 0 0 |
| − | 25 54 1 0 0 0 0 | + | 25 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 54 55 | + | M SAL 1 2 54 55 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 59 -6.8166 4.5057 | + | M SBV 1 59 -6.8166 4.5057 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AACGL0058 | + | ID FL7AACGL0058 |
| − | KNApSAcK_ID C00006829 | + | KNApSAcK_ID C00006829 |
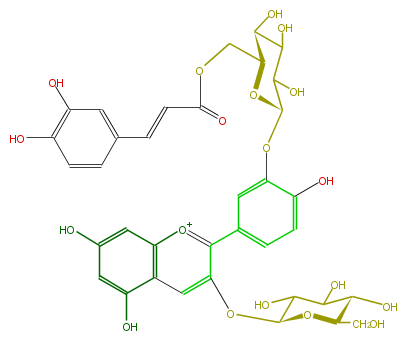
| − | NAME Cyanidin 3-glucoside-3'-(6''-caffeylglucoside) | + | NAME Cyanidin 3-glucoside-3'-(6''-caffeylglucoside) |
| − | CAS_RN 150070-20-5 | + | CAS_RN 150070-20-5 |
| − | FORMULA C36H37O19 | + | FORMULA C36H37O19 |
| − | EXACTMASS 773.192904002 | + | EXACTMASS 773.192904002 |
| − | AVERAGEMASS 773.66758 | + | AVERAGEMASS 773.66758 |
| − | SMILES OC(C6O)C(OC(C(O)6)COC(C=Cc(c5)ccc(c5O)O)=O)Oc(c1)c(ccc1c([o+1]3)c(cc(c(O)4)c(cc(c4)O)3)OC(C(O)2)OC(CO)C(O)C2O)O | + | SMILES OC(C6O)C(OC(C(O)6)COC(C=Cc(c5)ccc(c5O)O)=O)Oc(c1)c(ccc1c([o+1]3)c(cc(c(O)4)c(cc(c4)O)3)OC(C(O)2)OC(CO)C(O)C2O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-2.1465 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1465 -1.9810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5902 -2.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0339 -1.9810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0339 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5902 -1.0174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4775 -2.3022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0788 -1.9810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0788 -1.3386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4775 -1.0174 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
0.6348 -1.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2018 -1.3449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7688 -1.0176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7688 -0.3629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2018 -0.0355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6348 -0.3629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7026 -1.0176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3356 -0.0356 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5902 -2.9443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4948 -2.7017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2018 0.6190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7554 -2.3604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4546 -2.8814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0331 -2.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6150 -2.8814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9159 -2.3604 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3374 -2.5256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1967 -2.5101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5756 -2.1131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6192 -2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9532 1.6798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5211 1.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3409 1.9824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5211 2.6168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9532 2.9447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1333 2.3141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3155 3.3070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5211 3.0260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4696 2.7039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7585 1.7413 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1220 2.1738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1220 1.4250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3295 1.1643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8119 1.4250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1668 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7802 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1017 1.3672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7447 1.3672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0662 0.8104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7447 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1017 0.2535 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7084 0.8104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0658 1.9233 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9939 -2.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7084 -3.3070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
15 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 20 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
36 39 1 0 0 0 0
33 40 1 0 0 0 0
32 21 1 0 0 0 0
39 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
44 45 2 0 0 0 0
45 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 46 1 0 0 0 0
49 52 1 0 0 0 0
48 53 1 0 0 0 0
25 54 1 0 0 0 0
54 55 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 54 55
M SBL 1 1 59
M SMT 1 CH2OH
M SBV 1 59 -6.8166 4.5057
S SKP 8
ID FL7AACGL0058
KNApSAcK_ID C00006829
NAME Cyanidin 3-glucoside-3'-(6''-caffeylglucoside)
CAS_RN 150070-20-5
FORMULA C36H37O19
EXACTMASS 773.192904002
AVERAGEMASS 773.66758
SMILES OC(C6O)C(OC(C(O)6)COC(C=Cc(c5)ccc(c5O)O)=O)Oc(c1)c(ccc1c([o+1]3)c(cc(c(O)4)c(cc(c4)O)3)OC(C(O)2)OC(CO)C(O)C2O)O
M END