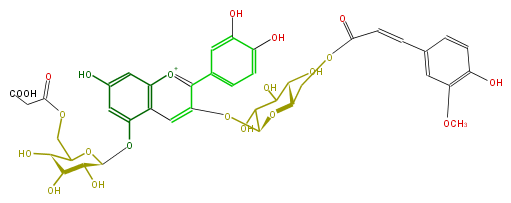
Mol:FL7AACGL0055
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 62 67 0 0 0 0 0 0 0 0999 V2000 | + | 62 67 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3429 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3429 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3429 0.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3429 0.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7866 -0.1431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7866 -0.1431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2303 0.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2303 0.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2303 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2303 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7866 1.1416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7866 1.1416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3260 -0.1431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3260 -0.1431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8823 0.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8823 0.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8823 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8823 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3260 1.1416 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.3260 1.1416 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4384 1.1415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4384 1.1415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0053 0.8141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0053 0.8141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5723 1.1415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5723 1.1415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5723 1.7961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5723 1.7961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0053 2.1235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0053 2.1235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4384 1.7961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4384 1.7961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8990 1.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8990 1.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1391 2.1234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1391 2.1234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7866 -0.7853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7866 -0.7853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8671 0.0044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8671 0.0044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0053 2.7780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0053 2.7780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9454 -1.0595 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.9454 -1.0595 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.5742 -1.5494 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5742 -1.5494 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.0397 -1.3416 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.0397 -1.3416 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.5240 -1.3360 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.5240 -1.3360 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.8988 -0.9611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8988 -0.9611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3664 -1.2079 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.3664 -1.2079 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.5363 -1.0077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5363 -1.0077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9246 -2.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9246 -2.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7335 -1.8557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7335 -1.8557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7370 -0.6328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7370 -0.6328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6292 0.1487 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.6292 0.1487 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.7338 -0.4438 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.7338 -0.4438 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.0706 0.0547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0706 0.0547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6227 0.3021 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.6227 0.3021 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.5182 0.8947 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.5182 0.8947 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.1813 0.3962 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.1813 0.3962 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.2975 -0.3251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2975 -0.3251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9081 0.7864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9081 0.7864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1782 1.2024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1782 1.2024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8659 0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8659 0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6152 0.0579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6152 0.0579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6162 1.5963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6162 1.5963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2507 2.0744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2507 2.0744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9513 2.3110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9513 2.3110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.6077 2.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.6077 2.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.2336 1.7441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.2336 1.7441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.2909 1.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.2909 1.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.8906 0.8054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.8906 0.8054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.4326 1.1849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.4326 1.1849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.3750 1.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.3750 1.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.7754 2.1236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.7754 2.1236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.0319 0.9054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.0319 0.9054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9816 2.6485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9816 2.6485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0963 0.4616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0963 0.4616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5740 0.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5740 0.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9745 0.6237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9745 0.6237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5230 0.4240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5230 0.4240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9874 1.0791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9874 1.0791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8732 1.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8732 1.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.8034 -0.1908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.8034 -0.1908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.7165 -1.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.7165 -1.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 25 19 1 0 0 0 0 | + | 25 19 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 37 39 1 0 0 0 0 | + | 37 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 31 42 1 0 0 0 0 | + | 31 42 1 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 44 54 2 0 0 0 0 | + | 44 54 2 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 42 55 1 0 0 0 0 | + | 42 55 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 56 57 1 0 0 0 0 | + | 56 57 1 0 0 0 0 |
| − | 57 58 2 0 0 0 0 | + | 57 58 2 0 0 0 0 |
| − | 55 59 2 0 0 0 0 | + | 55 59 2 0 0 0 0 |
| − | 57 60 1 0 0 0 0 | + | 57 60 1 0 0 0 0 |
| − | 49 61 1 0 0 0 0 | + | 49 61 1 0 0 0 0 |
| − | 61 62 1 0 0 0 0 | + | 61 62 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 61 62 | + | M SAL 2 2 61 62 |
| − | M SBL 2 1 66 | + | M SBL 2 1 66 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 66 1.1477 -0.481 | + | M SVB 2 66 1.1477 -0.481 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 57 58 60 | + | M SAL 1 3 57 58 60 |
| − | M SBL 1 1 62 | + | M SBL 1 1 62 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SVB 1 62 -4.2681 0.3166 | + | M SVB 1 62 -4.2681 0.3166 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AACGL0055 | + | ID FL7AACGL0055 |
| − | KNApSAcK_ID C00006826 | + | KNApSAcK_ID C00006826 |
| − | NAME Cyanidin 3-(6''-ferulylglucoside)-5-(6'''-malonylglucoside) | + | NAME Cyanidin 3-(6''-ferulylglucoside)-5-(6'''-malonylglucoside) |
| − | CAS_RN 123453-03-2 | + | CAS_RN 123453-03-2 |
| − | FORMULA C40H41O22 | + | FORMULA C40H41O22 |
| − | EXACTMASS 873.208947996 | + | EXACTMASS 873.208947996 |
| − | AVERAGEMASS 873.7403400000001 | + | AVERAGEMASS 873.7403400000001 |
| − | SMILES c(c6)(c(cc(c6)C=CC(OC[C@H]([C@H](O)5)O[C@H](C(O)C(O)5)Oc(c3c(c4)cc(c(O)c4)O)cc(c([o+1]3)1)c(O[C@H](O2)[C@H]([C@@H](O)[C@H](C(COC(=O)CC(O)=O)2)O)O)cc(O)c1)=O)OC)O | + | SMILES c(c6)(c(cc(c6)C=CC(OC[C@H]([C@H](O)5)O[C@H](C(O)C(O)5)Oc(c3c(c4)cc(c(O)c4)O)cc(c([o+1]3)1)c(O[C@H](O2)[C@H]([C@@H](O)[C@H](C(COC(=O)CC(O)=O)2)O)O)cc(O)c1)=O)OC)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
62 67 0 0 0 0 0 0 0 0999 V2000
-1.3429 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3429 0.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7866 -0.1431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2303 0.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2303 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7866 1.1416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3260 -0.1431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8823 0.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8823 0.8204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3260 1.1416 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.4384 1.1415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0053 0.8141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5723 1.1415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5723 1.7961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0053 2.1235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4384 1.7961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8990 1.1415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1391 2.1234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7866 -0.7853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8671 0.0044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0053 2.7780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9454 -1.0595 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.5742 -1.5494 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.0397 -1.3416 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.5240 -1.3360 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.8988 -0.9611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3664 -1.2079 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.5363 -1.0077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9246 -2.0107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7335 -1.8557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7370 -0.6328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6292 0.1487 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.7338 -0.4438 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.0706 0.0547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6227 0.3021 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.5182 0.8947 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.1813 0.3962 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.2975 -0.3251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9081 0.7864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1782 1.2024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8659 0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6152 0.0579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6162 1.5963 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2507 2.0744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9513 2.3110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.6077 2.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.2336 1.7441 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.2909 1.0851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.8906 0.8054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.4326 1.1849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.3750 1.8440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.7754 2.1236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.0319 0.9054 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9816 2.6485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0963 0.4616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5740 0.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9745 0.6237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5230 0.4240 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9874 1.0791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8732 1.1978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.8034 -0.1908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.7165 -1.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
15 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
27 31 1 0 0 0 0
25 19 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
37 39 1 0 0 0 0
36 40 1 0 0 0 0
35 41 1 0 0 0 0
33 20 1 0 0 0 0
31 42 1 0 0 0 0
41 43 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
44 45 1 0 0 0 0
44 54 2 0 0 0 0
43 44 1 0 0 0 0
42 55 1 0 0 0 0
55 56 1 0 0 0 0
56 57 1 0 0 0 0
57 58 2 0 0 0 0
55 59 2 0 0 0 0
57 60 1 0 0 0 0
49 61 1 0 0 0 0
61 62 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 61 62
M SBL 2 1 66
M SMT 2 OCH3
M SVB 2 66 1.1477 -0.481
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 57 58 60
M SBL 1 1 62
M SMT 1 COOH
M SVB 1 62 -4.2681 0.3166
S SKP 8
ID FL7AACGL0055
KNApSAcK_ID C00006826
NAME Cyanidin 3-(6''-ferulylglucoside)-5-(6'''-malonylglucoside)
CAS_RN 123453-03-2
FORMULA C40H41O22
EXACTMASS 873.208947996
AVERAGEMASS 873.7403400000001
SMILES c(c6)(c(cc(c6)C=CC(OC[C@H]([C@H](O)5)O[C@H](C(O)C(O)5)Oc(c3c(c4)cc(c(O)c4)O)cc(c([o+1]3)1)c(O[C@H](O2)[C@H]([C@@H](O)[C@H](C(COC(=O)CC(O)=O)2)O)O)cc(O)c1)=O)OC)O
M END