Mol:FL7AAAGL0055
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 79 86 0 0 0 0 0 0 0 0999 V2000 | + | 79 86 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1145 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1145 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1145 3.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1145 3.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4001 2.7945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4001 2.7945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3144 3.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3144 3.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3144 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3144 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4001 4.4445 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -0.4001 4.4445 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0289 4.4445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0289 4.4445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8290 4.4445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8290 4.4445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5435 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5435 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5435 3.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5435 3.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8290 2.7945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8290 2.7945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7433 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7433 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4578 4.4445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4578 4.4445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4578 5.2695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4578 5.2695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7433 5.6820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7433 5.6820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0289 5.2695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0289 5.2695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1723 5.6820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1723 5.6820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2580 4.4445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2580 4.4445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4153 0.2999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4153 0.2999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4153 1.1249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4153 1.1249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6196 1.3429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6196 1.3429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0372 1.9206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0372 1.9206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0371 1.0956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0371 1.0956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8328 0.8775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8328 0.8775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8328 0.0525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8328 0.0525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6196 2.1679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6196 2.1679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2122 0.0864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2122 0.0864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9986 0.5415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9986 0.5415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6297 2.4704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6297 2.4704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2172 1.7560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2172 1.7560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4191 1.9651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4191 1.9651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6259 1.7560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6259 1.7560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0383 2.4704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0383 2.4704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8365 2.2614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8365 2.2614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2490 2.9759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2490 2.9759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8290 1.9695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8290 1.9695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0066 1.2506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0066 1.2506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4266 2.2569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4266 2.2569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0141 1.9695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0141 1.9695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0459 3.1894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0459 3.1894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9954 0.1185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9954 0.1185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4079 0.8330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4079 0.8330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8278 1.4196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8278 1.4196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6122 2.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6122 2.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1997 1.4967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1997 1.4967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7798 0.9100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7798 0.9100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3673 0.1955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3673 0.1955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0289 2.7945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0289 2.7945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2403 2.1341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2403 2.1341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5788 -0.4648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5788 -0.4648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6214 0.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6214 0.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5808 -0.6014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5808 -0.6014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1683 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1683 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3433 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3433 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5808 -2.0303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5808 -2.0303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0692 -2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0692 -2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8942 -2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8942 -2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6297 -1.3277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6297 -1.3277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4266 -1.5412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4266 -1.5412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0464 -1.9111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0464 -1.9111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2599 -2.7080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2599 -2.7080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6765 -3.2913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6765 -3.2913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8796 -3.0778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8796 -3.0778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2963 -3.6612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2963 -3.6612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5098 -4.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5098 -4.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3067 -4.6716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3067 -4.6716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8900 -4.0882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8900 -4.0882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3067 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3067 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1317 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1317 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5442 -2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5442 -2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1317 -2.7448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1317 -2.7448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3067 -2.7448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3067 -2.7448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3692 -2.0303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3692 -2.0303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5442 -3.4592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5442 -3.4592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1317 -4.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1317 -4.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9264 -5.0414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9264 -5.0414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5202 -5.4685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5202 -5.4685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3171 -5.6820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3171 -5.6820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4162 -0.5308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4162 -0.5308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 5 7 1 0 0 0 0 | + | 5 7 1 0 0 0 0 |
| − | 1 8 1 0 0 0 0 | + | 1 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 2 1 0 0 0 0 | + | 11 2 1 0 0 0 0 |
| − | 7 12 2 0 0 0 0 | + | 7 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 7 1 0 0 0 0 | + | 16 7 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 11 36 1 0 0 0 0 | + | 11 36 1 0 0 0 0 |
| − | 9 18 1 0 0 0 0 | + | 9 18 1 0 0 0 0 |
| − | 4 48 1 0 0 0 0 | + | 4 48 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 22 49 1 0 0 0 0 | + | 22 49 1 0 0 0 0 |
| − | 21 26 1 0 0 0 0 | + | 21 26 1 0 0 0 0 |
| − | 19 27 1 0 0 0 0 | + | 19 27 1 0 0 0 0 |
| − | 20 28 1 0 0 0 0 | + | 20 28 1 0 0 0 0 |
| − | 25 79 1 0 0 0 0 | + | 25 79 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 29 38 1 0 0 0 0 | + | 29 38 1 0 0 0 0 |
| − | 30 39 1 0 0 0 0 | + | 30 39 1 0 0 0 0 |
| − | 35 40 1 0 0 0 0 | + | 35 40 1 0 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 44 48 1 0 0 0 0 | + | 44 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 41 50 1 0 0 0 0 | + | 41 50 1 0 0 0 0 |
| − | 42 51 1 0 0 0 0 | + | 42 51 1 0 0 0 0 |
| − | 47 52 1 0 0 0 0 | + | 47 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 53 55 2 0 0 0 0 | + | 53 55 2 0 0 0 0 |
| − | 54 56 2 0 0 0 0 | + | 54 56 2 0 0 0 0 |
| − | 56 57 1 0 0 0 0 | + | 56 57 1 0 0 0 0 |
| − | 58 59 2 0 0 0 0 | + | 58 59 2 0 0 0 0 |
| − | 58 60 1 0 0 0 0 | + | 58 60 1 0 0 0 0 |
| − | 60 61 2 0 0 0 0 | + | 60 61 2 0 0 0 0 |
| − | 61 62 1 0 0 0 0 | + | 61 62 1 0 0 0 0 |
| − | 62 63 2 0 0 0 0 | + | 62 63 2 0 0 0 0 |
| − | 63 64 1 0 0 0 0 | + | 63 64 1 0 0 0 0 |
| − | 64 65 2 0 0 0 0 | + | 64 65 2 0 0 0 0 |
| − | 65 66 1 0 0 0 0 | + | 65 66 1 0 0 0 0 |
| − | 66 67 2 0 0 0 0 | + | 66 67 2 0 0 0 0 |
| − | 67 62 1 0 0 0 0 | + | 67 62 1 0 0 0 0 |
| − | 57 68 2 0 0 0 0 | + | 57 68 2 0 0 0 0 |
| − | 68 69 1 0 0 0 0 | + | 68 69 1 0 0 0 0 |
| − | 69 70 2 0 0 0 0 | + | 69 70 2 0 0 0 0 |
| − | 70 71 1 0 0 0 0 | + | 70 71 1 0 0 0 0 |
| − | 71 72 2 0 0 0 0 | + | 71 72 2 0 0 0 0 |
| − | 72 57 1 0 0 0 0 | + | 72 57 1 0 0 0 0 |
| − | 70 73 1 0 0 0 0 | + | 70 73 1 0 0 0 0 |
| − | 71 74 1 0 0 0 0 | + | 71 74 1 0 0 0 0 |
| − | 74 75 1 0 0 0 0 | + | 74 75 1 0 0 0 0 |
| − | 65 76 1 0 0 0 0 | + | 65 76 1 0 0 0 0 |
| − | 66 77 1 0 0 0 0 | + | 66 77 1 0 0 0 0 |
| − | 77 78 1 0 0 0 0 | + | 77 78 1 0 0 0 0 |
| − | 58 79 1 0 0 0 0 | + | 58 79 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAAGL0055 | + | ID FL7AAAGL0055 |
| − | KNApSAcK_ID C00014025 | + | KNApSAcK_ID C00014025 |
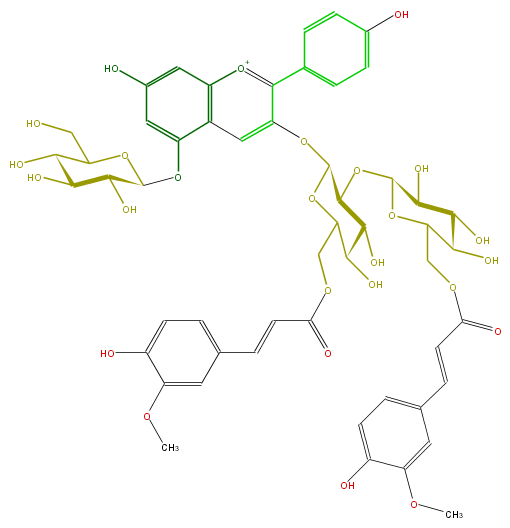
| − | NAME Pelargonidin 3-O-[2-O-(6-(E)-feruloyl-beta-D-glucopyranosyl)-6-O-(E)-feruloyl-beta-D-glucopyranoside]-5-O-(beta-D-glucopyranoside);Pelargonidin 3-(2-(6-ferulylglucosyl)-6-ferulylglucoside)-5-glucoside | + | NAME Pelargonidin 3-O-[2-O-(6-(E)-feruloyl-beta-D-glucopyranosyl)-6-O-(E)-feruloyl-beta-D-glucopyranoside]-5-O-(beta-D-glucopyranoside);Pelargonidin 3-(2-(6-ferulylglucosyl)-6-ferulylglucoside)-5-glucoside |
| − | CAS_RN 185027-89-8 | + | CAS_RN 185027-89-8 |
| − | FORMULA C53H57O26 | + | FORMULA C53H57O26 |
| − | EXACTMASS 1109.313806996 | + | EXACTMASS 1109.313806996 |
| − | AVERAGEMASS 1110.00408 | + | AVERAGEMASS 1110.00408 |
| − | SMILES c(c1)(ccc(c([o+1]4)c(OC(O5)C(OC(C7O)OC(COC(=O)C=Cc(c8)cc(c(O)c8)OC)C(O)C7O)C(C(C(COC(=O)C=Cc(c6)ccc(O)c(OC)6)5)O)O)cc(c24)c(OC(C3O)OC(C(C(O)3)O)CO)cc(c2)O)c1)O | + | SMILES c(c1)(ccc(c([o+1]4)c(OC(O5)C(OC(C7O)OC(COC(=O)C=Cc(c8)cc(c(O)c8)OC)C(O)C7O)C(C(C(COC(=O)C=Cc(c6)ccc(O)c(OC)6)5)O)O)cc(c24)c(OC(C3O)OC(C(C(O)3)O)CO)cc(c2)O)c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
79 86 0 0 0 0 0 0 0 0999 V2000
-1.1145 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1145 3.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4001 2.7945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3144 3.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3144 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4001 4.4445 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
1.0289 4.4445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8290 4.4445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5435 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5435 3.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8290 2.7945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7433 4.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4578 4.4445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4578 5.2695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7433 5.6820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0289 5.2695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1723 5.6820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2580 4.4445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4153 0.2999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4153 1.1249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6196 1.3429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0372 1.9206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0371 1.0956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8328 0.8775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8328 0.0525 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6196 2.1679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2122 0.0864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9986 0.5415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6297 2.4704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2172 1.7560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4191 1.9651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6259 1.7560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0383 2.4704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8365 2.2614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2490 2.9759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8290 1.9695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0066 1.2506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4266 2.2569 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0141 1.9695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0459 3.1894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9954 0.1185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4079 0.8330 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8278 1.4196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6122 2.2111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1997 1.4967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7798 0.9100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3673 0.1955 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0289 2.7945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2403 2.1341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5788 -0.4648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6214 0.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5808 -0.6014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1683 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3433 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5808 -2.0303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0692 -2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8942 -2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6297 -1.3277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4266 -1.5412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0464 -1.9111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2599 -2.7080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6765 -3.2913 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8796 -3.0778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2963 -3.6612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5098 -4.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3067 -4.6716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8900 -4.0882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3067 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1317 -1.3158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5442 -2.0303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1317 -2.7448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3067 -2.7448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3692 -2.0303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5442 -3.4592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1317 -4.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9264 -5.0414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5202 -5.4685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3171 -5.6820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4162 -0.5308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
5 7 1 0 0 0 0
1 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 2 1 0 0 0 0
7 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 7 1 0 0 0 0
14 17 1 0 0 0 0
11 36 1 0 0 0 0
9 18 1 0 0 0 0
4 48 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
24 25 1 0 0 0 0
22 49 1 0 0 0 0
21 26 1 0 0 0 0
19 27 1 0 0 0 0
20 28 1 0 0 0 0
25 79 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
34 35 1 0 0 0 0
32 36 1 0 0 0 0
31 37 1 0 0 0 0
29 38 1 0 0 0 0
30 39 1 0 0 0 0
35 40 1 0 0 0 0
41 42 1 1 0 0 0
42 43 1 1 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
46 47 1 0 0 0 0
44 48 1 0 0 0 0
43 49 1 0 0 0 0
41 50 1 0 0 0 0
42 51 1 0 0 0 0
47 52 1 0 0 0 0
52 53 1 0 0 0 0
53 54 1 0 0 0 0
53 55 2 0 0 0 0
54 56 2 0 0 0 0
56 57 1 0 0 0 0
58 59 2 0 0 0 0
58 60 1 0 0 0 0
60 61 2 0 0 0 0
61 62 1 0 0 0 0
62 63 2 0 0 0 0
63 64 1 0 0 0 0
64 65 2 0 0 0 0
65 66 1 0 0 0 0
66 67 2 0 0 0 0
67 62 1 0 0 0 0
57 68 2 0 0 0 0
68 69 1 0 0 0 0
69 70 2 0 0 0 0
70 71 1 0 0 0 0
71 72 2 0 0 0 0
72 57 1 0 0 0 0
70 73 1 0 0 0 0
71 74 1 0 0 0 0
74 75 1 0 0 0 0
65 76 1 0 0 0 0
66 77 1 0 0 0 0
77 78 1 0 0 0 0
58 79 1 0 0 0 0
S SKP 8
ID FL7AAAGL0055
KNApSAcK_ID C00014025
NAME Pelargonidin 3-O-[2-O-(6-(E)-feruloyl-beta-D-glucopyranosyl)-6-O-(E)-feruloyl-beta-D-glucopyranoside]-5-O-(beta-D-glucopyranoside);Pelargonidin 3-(2-(6-ferulylglucosyl)-6-ferulylglucoside)-5-glucoside
CAS_RN 185027-89-8
FORMULA C53H57O26
EXACTMASS 1109.313806996
AVERAGEMASS 1110.00408
SMILES c(c1)(ccc(c([o+1]4)c(OC(O5)C(OC(C7O)OC(COC(=O)C=Cc(c8)cc(c(O)c8)OC)C(O)C7O)C(C(C(COC(=O)C=Cc(c6)ccc(O)c(OC)6)5)O)O)cc(c24)c(OC(C3O)OC(C(C(O)3)O)CO)cc(c2)O)c1)O
M END