Mol:FL7AAAGL0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.0494 2.0611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0494 2.0611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0494 1.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0494 1.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4931 1.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4931 1.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9368 1.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9368 1.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9368 2.0611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9368 2.0611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4931 2.3823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4931 2.3823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3805 1.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3805 1.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8242 1.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8242 1.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8242 2.0611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8242 2.0611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3805 2.3823 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -1.3805 2.3823 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2681 2.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2681 2.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2989 2.0548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2989 2.0548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8659 2.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8659 2.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8659 3.0369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8659 3.0369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2989 3.3642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2989 3.3642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2681 3.0369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2681 3.0369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4327 3.3641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4327 3.3641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6055 2.3822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6055 2.3822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4931 0.4554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4931 0.4554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0683 0.5876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0683 0.5876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2200 1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2200 1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8323 0.5234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8323 0.5234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5778 0.7365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5778 0.7365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3278 0.5234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3278 0.5234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7155 1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7155 1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9700 0.9819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9700 0.9819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5000 1.0019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5000 1.0019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2769 1.5135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2769 1.5135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2749 0.8719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2749 0.8719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0385 0.5234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0385 0.5234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9097 -0.0544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9097 -0.0544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8058 -2.2635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8058 -2.2635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6531 -2.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6531 -2.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7743 -1.3699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7743 -1.3699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1258 -0.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1258 -0.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4145 -0.9360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4145 -0.9360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3866 -1.6700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3866 -1.6700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8058 -2.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8058 -2.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4026 -3.0919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4026 -3.0919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6055 -1.3585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6055 -1.3585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4823 -1.9123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4823 -1.9123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6032 -2.2581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6032 -2.2581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0876 -2.9388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0876 -2.9388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3451 -2.6500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3451 -2.6500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3714 -2.6423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3714 -2.6423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1492 -2.1215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1492 -2.1215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9077 -2.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9077 -2.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1797 -2.5910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1797 -2.5910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8914 -2.9779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8914 -2.9779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0804 -3.3642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0804 -3.3642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5843 -2.2735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5843 -2.2735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2032 -1.5865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2032 -1.5865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4888 -1.9990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4888 -1.9990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 42 1 0 0 0 0 | + | 47 42 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 51 41 1 0 0 0 0 | + | 51 41 1 0 0 0 0 |
| − | 47 52 1 0 0 0 0 | + | 47 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 57 -7.5483 5.6735 | + | M SBV 1 57 -7.5483 5.6735 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAAGL0008 | + | ID FL7AAAGL0008 |
| − | KNApSAcK_ID C00006642 | + | KNApSAcK_ID C00006642 |
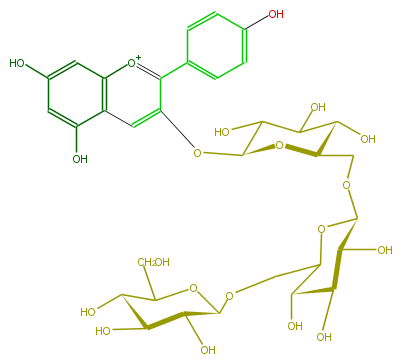
| − | NAME Pelargonidin 3-gentiotrioside | + | NAME Pelargonidin 3-gentiotrioside |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C33H41O20 | + | FORMULA C33H41O20 |
| − | EXACTMASS 757.219118752 | + | EXACTMASS 757.219118752 |
| − | AVERAGEMASS 757.6666399999999 | + | AVERAGEMASS 757.6666399999999 |
| − | SMILES C(C(O)1)(C(C(OC1OCC(C6O)OC(C(C6O)O)OCC(C5O)OC(C(C(O)5)O)Oc(c3c(c4)ccc(c4)O)cc(c([o+1]3)2)c(cc(O)c2)O)CO)O)O | + | SMILES C(C(O)1)(C(C(OC1OCC(C6O)OC(C(C6O)O)OCC(C5O)OC(C(C(O)5)O)Oc(c3c(c4)ccc(c4)O)cc(c([o+1]3)2)c(cc(O)c2)O)CO)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-3.0494 2.0611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0494 1.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4931 1.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9368 1.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9368 2.0611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4931 2.3823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3805 1.0976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8242 1.4187 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8242 2.0611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3805 2.3823 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
-0.2681 2.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2989 2.0548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8659 2.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8659 3.0369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2989 3.3642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2681 3.0369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4327 3.3641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6055 2.3822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4931 0.4554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0683 0.5876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2200 1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8323 0.5234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5778 0.7365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3278 0.5234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7155 1.1949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9700 0.9819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5000 1.0019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2769 1.5135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2749 0.8719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0385 0.5234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9097 -0.0544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8058 -2.2635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6531 -2.1573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7743 -1.3699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1258 -0.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4145 -0.9360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3866 -1.6700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8058 -2.8759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4026 -3.0919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6055 -1.3585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4823 -1.9123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6032 -2.2581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0876 -2.9388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3451 -2.6500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3714 -2.6423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1492 -2.1215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9077 -2.3939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1797 -2.5910 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8914 -2.9779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0804 -3.3642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5843 -2.2735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2032 -1.5865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4888 -1.9990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
8 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
30 31 1 0 0 0 0
22 20 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
37 41 1 0 0 0 0
31 35 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
51 41 1 0 0 0 0
47 52 1 0 0 0 0
52 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 57
M SMT 1 ^CH2OH
M SBV 1 57 -7.5483 5.6735
S SKP 8
ID FL7AAAGL0008
KNApSAcK_ID C00006642
NAME Pelargonidin 3-gentiotrioside
CAS_RN -
FORMULA C33H41O20
EXACTMASS 757.219118752
AVERAGEMASS 757.6666399999999
SMILES C(C(O)1)(C(C(OC1OCC(C6O)OC(C(C6O)O)OCC(C5O)OC(C(C(O)5)O)Oc(c3c(c4)ccc(c4)O)cc(c([o+1]3)2)c(cc(O)c2)O)CO)O)O
M END