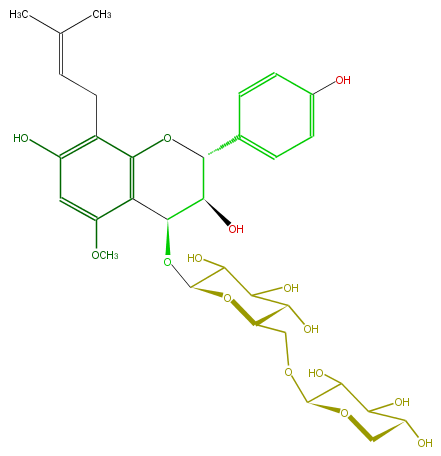
Mol:FL6DBAGI0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 47 51 0 0 0 0 0 0 0 0999 V2000 | + | 47 51 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.2555 1.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2555 1.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2555 0.5724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2555 0.5724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5408 0.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5408 0.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8262 0.5724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8262 0.5724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8262 1.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8262 1.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5408 1.8103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5408 1.8103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1115 0.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1115 0.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3969 0.5724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3969 0.5724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3969 1.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3969 1.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1115 1.8103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1115 1.8103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3175 1.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3175 1.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0459 1.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0459 1.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7742 1.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7742 1.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7742 2.6512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7742 2.6512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0459 3.0717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0459 3.0717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3175 2.6512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3175 2.6512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1115 -0.6651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1115 -0.6651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1991 -0.0236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1991 -0.0236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5408 2.6354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5408 2.6354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2555 3.0480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2555 3.0480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9701 1.8103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9701 1.8103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2555 3.8730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2555 3.8730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9699 4.2854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9699 4.2854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5411 4.2854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5411 4.2854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0127 -0.8114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0127 -0.8114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6567 -1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6567 -1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0931 -1.3854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0931 -1.3854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6343 -1.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6343 -1.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3038 -1.5567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3038 -1.5567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5540 -1.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5540 -1.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4974 -0.5998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4974 -0.5998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2916 -1.1882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2916 -1.1882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6847 -2.0202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6847 -2.0202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2494 -2.1132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2494 -2.1132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3558 -3.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3558 -3.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6863 -3.5118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6863 -3.5118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4362 -3.6995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4362 -3.6995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9774 -4.2572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9774 -4.2572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6468 -3.8709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6468 -3.8709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8971 -3.6830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8971 -3.6830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9309 -2.8959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9309 -2.8959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5109 -3.4815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5109 -3.4815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9701 -4.2854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9701 -4.2854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3065 -2.8713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3065 -2.8713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3288 2.9713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3288 2.9713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5482 -0.5431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5482 -0.5431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6303 -1.0730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6303 -1.0730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 6 0 0 0 | + | 9 11 1 6 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 1 1 0 0 0 | + | 7 17 1 1 0 0 0 |
| − | 8 18 1 1 0 0 0 | + | 8 18 1 1 0 0 0 |
| − | 6 19 1 0 0 0 0 | + | 6 19 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 21 1 1 0 0 0 0 | + | 21 1 1 0 0 0 0 |
| − | 20 22 2 0 0 0 0 | + | 20 22 2 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 22 24 1 0 0 0 0 | + | 22 24 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 29 28 1 1 0 0 0 | + | 29 28 1 1 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 25 1 0 0 0 0 | + | 30 25 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 29 33 1 0 0 0 0 | + | 29 33 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 39 38 1 1 0 0 0 | + | 39 38 1 1 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 26 17 1 0 0 0 0 | + | 26 17 1 0 0 0 0 |
| − | 34 44 1 0 0 0 0 | + | 34 44 1 0 0 0 0 |
| − | 44 36 1 0 0 0 0 | + | 44 36 1 0 0 0 0 |
| − | 14 45 1 0 0 0 0 | + | 14 45 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 3 46 1 0 0 0 0 | + | 3 46 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 46 47 | + | M SAL 1 2 46 47 |
| − | M SBL 1 1 51 | + | M SBL 1 1 51 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 51 0.0074 0.7030 | + | M SBV 1 51 0.0074 0.7030 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL6DBAGI0002 | + | ID FL6DBAGI0002 |
| − | FORMULA C32H42O15 | + | FORMULA C32H42O15 |
| − | EXACTMASS 666.252370674 | + | EXACTMASS 666.252370674 |
| − | AVERAGEMASS 666.66688 | + | AVERAGEMASS 666.66688 |
| − | SMILES c(c5OC)(c4c(CC=C(C)C)c(O)c5)C(C(C(O4)c(c3)ccc(c3)O)O)OC(O1)C(C(O)C(O)C1COC(O2)C(O)C(C(C2)O)O)O | + | SMILES c(c5OC)(c4c(CC=C(C)C)c(O)c5)C(C(C(O4)c(c3)ccc(c3)O)O)OC(O1)C(C(O)C(O)C1COC(O2)C(O)C(C(C2)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
47 51 0 0 0 0 0 0 0 0999 V2000
-3.2555 1.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2555 0.5724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5408 0.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8262 0.5724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8262 1.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5408 1.8103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1115 0.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3969 0.5724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3969 1.3976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1115 1.8103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3175 1.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0459 1.3895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7742 1.8101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7742 2.6512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0459 3.0717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3175 2.6512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1115 -0.6651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1991 -0.0236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5408 2.6354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2555 3.0480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9701 1.8103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2555 3.8730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9699 4.2854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5411 4.2854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0127 -0.8114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6567 -1.1978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0931 -1.3854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6343 -1.9432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3038 -1.5567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5540 -1.3691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4974 -0.5998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2916 -1.1882 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6847 -2.0202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2494 -2.1132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3558 -3.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6863 -3.5118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4362 -3.6995 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9774 -4.2572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6468 -3.8709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8971 -3.6830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9309 -2.8959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5109 -3.4815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9701 -4.2854 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3065 -2.8713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3288 2.9713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5482 -0.5431 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6303 -1.0730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 6 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 1 1 0 0 0
8 18 1 1 0 0 0
6 19 1 0 0 0 0
19 20 1 0 0 0 0
21 1 1 0 0 0 0
20 22 2 0 0 0 0
22 23 1 0 0 0 0
22 24 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 28 1 1 0 0 0
29 28 1 1 0 0 0
29 30 1 0 0 0 0
30 25 1 0 0 0 0
25 31 1 0 0 0 0
30 32 1 0 0 0 0
29 33 1 0 0 0 0
28 34 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 38 1 1 0 0 0
39 38 1 1 0 0 0
39 40 1 0 0 0 0
40 35 1 0 0 0 0
35 41 1 0 0 0 0
40 42 1 0 0 0 0
39 43 1 0 0 0 0
26 17 1 0 0 0 0
34 44 1 0 0 0 0
44 36 1 0 0 0 0
14 45 1 0 0 0 0
46 47 1 0 0 0 0
3 46 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 46 47
M SBL 1 1 51
M SMT 1 OCH3
M SBV 1 51 0.0074 0.7030
S SKP 5
ID FL6DBAGI0002
FORMULA C32H42O15
EXACTMASS 666.252370674
AVERAGEMASS 666.66688
SMILES c(c5OC)(c4c(CC=C(C)C)c(O)c5)C(C(C(O4)c(c3)ccc(c3)O)O)OC(O1)C(C(O)C(O)C1COC(O2)C(O)C(C(C2)O)O)O
M END