Mol:FL6DAGGS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.2145 0.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2145 0.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2145 0.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2145 0.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6582 -0.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6582 -0.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1019 0.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1019 0.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1019 0.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1019 0.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6582 1.2451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6582 1.2451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5456 -0.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5456 -0.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9893 0.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9893 0.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9893 0.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9893 0.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5456 1.2451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5456 1.2451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4332 1.2450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4332 1.2450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1337 0.9176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1337 0.9176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7007 1.2450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7007 1.2450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7007 1.8997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7007 1.8997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1337 2.2270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1337 2.2270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4332 1.8997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4332 1.8997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5456 -0.6817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5456 -0.6817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7694 1.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7694 1.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2675 2.2269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2675 2.2269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6582 -0.7470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6582 -0.7470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5254 -0.1823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5254 -0.1823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1337 2.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1337 2.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4411 -1.0667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4411 -1.0667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9765 -1.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9765 -1.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8066 -0.7814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8066 -0.7814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9765 -0.1833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9765 -0.1833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4411 0.1258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4411 0.1258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6109 -0.4686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6109 -0.4686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2674 -0.4686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2674 -0.4686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5786 -1.5800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5786 -1.5800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7419 -2.2511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7419 -2.2511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3352 -1.0866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3352 -1.0866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2675 0.9177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2675 0.9177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8423 -1.9747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8423 -1.9747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5415 -2.4957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5415 -2.4957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1200 -2.3304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1200 -2.3304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7020 -2.4957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7020 -2.4957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0028 -1.9747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0028 -1.9747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4243 -2.1400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4243 -2.1400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2836 -2.1244 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2836 -2.1244 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6625 -1.7274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6625 -1.7274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4369 -2.2253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4369 -2.2253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0549 -2.4690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0549 -2.4690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7694 -2.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7694 -2.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 1 0 0 0 0 | + | 7 17 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 15 22 1 0 0 0 0 | + | 15 22 1 0 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 23 1 1 0 0 0 | + | 28 23 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 27 21 1 0 0 0 0 | + | 27 21 1 0 0 0 0 |
| − | 13 33 1 0 0 0 0 | + | 13 33 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 38 37 1 1 0 0 0 | + | 38 37 1 1 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 37 43 1 0 0 0 0 | + | 37 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 47 -4.7756 5.0344 | + | M SBV 1 47 -4.7756 5.0344 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL6DAGGS0003 | + | ID FL6DAGGS0003 |
| − | KNApSAcK_ID C00009022 | + | KNApSAcK_ID C00009022 |
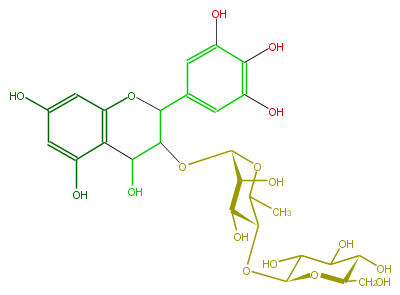
| − | NAME Leucodelphinidin 3-O-(beta-D-glucopyranosyl-(1->4)-alpha-L-rhamnopyranoside) | + | NAME Leucodelphinidin 3-O-(beta-D-glucopyranosyl-(1->4)-alpha-L-rhamnopyranoside) |
| − | CAS_RN 76520-51-9 | + | CAS_RN 76520-51-9 |
| − | FORMULA C27H34O17 | + | FORMULA C27H34O17 |
| − | EXACTMASS 630.179599662 | + | EXACTMASS 630.179599662 |
| − | AVERAGEMASS 630.54866 | + | AVERAGEMASS 630.54866 |
| − | SMILES C(C)(O1)C(OC(O5)C(O)C(O)C(C5CO)O)C(C(O)C(OC(C(c(c4)cc(c(c(O)4)O)O)2)C(O)c(c3O)c(cc(c3)O)O2)1)O | + | SMILES C(C)(O1)C(OC(O5)C(O)C(O)C(C5CO)O)C(C(O)C(OC(C(c(c4)cc(c(c(O)4)O)O)2)C(O)c(c3O)c(cc(c3)O)O2)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-3.2145 0.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2145 0.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6582 -0.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1019 0.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1019 0.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6582 1.2451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5456 -0.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9893 0.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9893 0.9239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5456 1.2451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4332 1.2450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1337 0.9176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7007 1.2450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7007 1.8997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1337 2.2270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4332 1.8997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5456 -0.6817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7694 1.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2675 2.2269 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6582 -0.7470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5254 -0.1823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1337 2.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4411 -1.0667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9765 -1.3758 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8066 -0.7814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9765 -0.1833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4411 0.1258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6109 -0.4686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2674 -0.4686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5786 -1.5800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7419 -2.2511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3352 -1.0866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2675 0.9177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8423 -1.9747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5415 -2.4957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1200 -2.3304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7020 -2.4957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0028 -1.9747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4243 -2.1400 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2836 -2.1244 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6625 -1.7274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4369 -2.2253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0549 -2.4690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7694 -2.8815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 1 0 0 0 0
18 1 1 0 0 0 0
14 19 1 0 0 0 0
3 20 1 0 0 0 0
8 21 1 0 0 0 0
15 22 1 0 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 1 0 0 0
28 23 1 1 0 0 0
28 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
25 32 1 0 0 0 0
27 21 1 0 0 0 0
13 33 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 37 1 1 0 0 0
38 37 1 1 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
34 40 1 0 0 0 0
39 41 1 0 0 0 0
38 42 1 0 0 0 0
35 31 1 0 0 0 0
37 43 1 0 0 0 0
43 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 47
M SMT 1 CH2OH
M SBV 1 47 -4.7756 5.0344
S SKP 8
ID FL6DAGGS0003
KNApSAcK_ID C00009022
NAME Leucodelphinidin 3-O-(beta-D-glucopyranosyl-(1->4)-alpha-L-rhamnopyranoside)
CAS_RN 76520-51-9
FORMULA C27H34O17
EXACTMASS 630.179599662
AVERAGEMASS 630.54866
SMILES C(C)(O1)C(OC(O5)C(O)C(O)C(C5CO)O)C(C(O)C(OC(C(c(c4)cc(c(c(O)4)O)O)2)C(O)c(c3O)c(cc(c3)O)O2)1)O
M END