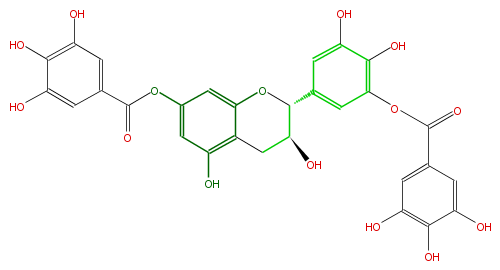
Mol:FL63AGNS0013
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3787 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3787 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3787 -0.0504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3787 -0.0504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8401 -0.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8401 -0.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3016 -0.0504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3016 -0.0504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3016 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3016 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8401 0.8823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8401 0.8823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2369 -0.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2369 -0.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7754 -0.0504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7754 -0.0504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7754 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7754 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2369 0.8823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2369 0.8823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9169 0.8822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9169 0.8822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2550 0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2550 0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7997 0.5338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7997 0.5338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3444 0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3444 0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3444 1.4772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3444 1.4772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7997 1.7917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7997 1.7917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2550 1.4772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2550 1.4772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8401 -0.9828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8401 -0.9828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7997 2.4201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7997 2.4201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1973 -0.6003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1973 -0.6003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8886 1.7914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8886 1.7914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8886 0.5341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8886 0.5341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4551 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4551 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9934 0.8822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9934 0.8822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4551 -0.0501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4551 -0.0501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9934 1.4996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9934 1.4996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5281 1.8083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5281 1.8083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0627 1.4996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0627 1.4996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0627 0.8822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0627 0.8822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5281 0.5735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5281 0.5735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5281 2.4256 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5281 2.4256 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5974 1.8082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5974 1.8082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5974 0.5735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5974 0.5735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5566 0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5566 0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1395 0.4849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1395 0.4849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5566 -0.6230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5566 -0.6230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0363 -0.9234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0363 -0.9234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0363 -1.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0363 -1.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5566 -1.8248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5566 -1.8248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0770 -1.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0770 -1.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0770 -0.9234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0770 -0.9234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5159 -1.8248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5159 -1.8248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5566 -2.4256 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5566 -2.4256 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5974 -1.8248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5974 -1.8248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 1 11 1 0 0 0 0 | + | 1 11 1 0 0 0 0 |
| − | 9 12 1 6 0 0 0 | + | 9 12 1 6 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 8 20 1 1 0 0 0 | + | 8 20 1 1 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 14 22 1 0 0 0 0 | + | 14 22 1 0 0 0 0 |
| − | 11 23 1 0 0 0 0 | + | 11 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 23 25 2 0 0 0 0 | + | 23 25 2 0 0 0 0 |
| − | 24 26 2 0 0 0 0 | + | 24 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 2 0 0 0 0 | + | 29 30 2 0 0 0 0 |
| − | 30 24 1 0 0 0 0 | + | 30 24 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 29 33 1 0 0 0 0 | + | 29 33 1 0 0 0 0 |
| − | 22 34 1 0 0 0 0 | + | 22 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 40 44 1 0 0 0 0 | + | 40 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL63AGNS0013 | + | ID FL63AGNS0013 |
| − | KNApSAcK_ID C00008891 | + | KNApSAcK_ID C00008891 |
| − | NAME Gallocatechin 7,3'-di-O-gallate | + | NAME Gallocatechin 7,3'-di-O-gallate |
| − | CAS_RN 142784-35-8 | + | CAS_RN 142784-35-8 |
| − | FORMULA C29H22O15 | + | FORMULA C29H22O15 |
| − | EXACTMASS 610.095870034 | + | EXACTMASS 610.095870034 |
| − | AVERAGEMASS 610.4759799999999 | + | AVERAGEMASS 610.4759799999999 |
| − | SMILES C(c(c5)cc(c(O)c5O)O)(Oc(c4)cc(c(c(O)4)3)OC(C(C3)O)c(c2)cc(c(O)c2O)OC(c(c1)cc(c(c1O)O)O)=O)=O | + | SMILES C(c(c5)cc(c(O)c5O)O)(Oc(c4)cc(c(c(O)4)3)OC(C(C3)O)c(c2)cc(c(O)c2O)OC(c(c1)cc(c(c1O)O)O)=O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.3787 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3787 -0.0504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8401 -0.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3016 -0.0504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3016 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8401 0.8823 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2369 -0.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7754 -0.0504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7754 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2369 0.8823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9169 0.8822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2550 0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7997 0.5338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3444 0.8483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3444 1.4772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7997 1.7917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2550 1.4772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8401 -0.9828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7997 2.4201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1973 -0.6003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8886 1.7914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8886 0.5341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4551 0.5714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9934 0.8822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4551 -0.0501 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9934 1.4996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5281 1.8083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0627 1.4996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0627 0.8822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5281 0.5735 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5281 2.4256 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5974 1.8082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5974 0.5735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5566 0.1484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1395 0.4849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5566 -0.6230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0363 -0.9234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0363 -1.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5566 -1.8248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0770 -1.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0770 -0.9234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5159 -1.8248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5566 -2.4256 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5974 -1.8248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
1 11 1 0 0 0 0
9 12 1 6 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
16 19 1 0 0 0 0
8 20 1 1 0 0 0
15 21 1 0 0 0 0
14 22 1 0 0 0 0
11 23 1 0 0 0 0
23 24 1 0 0 0 0
23 25 2 0 0 0 0
24 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 29 1 0 0 0 0
29 30 2 0 0 0 0
30 24 1 0 0 0 0
27 31 1 0 0 0 0
28 32 1 0 0 0 0
29 33 1 0 0 0 0
22 34 1 0 0 0 0
34 35 2 0 0 0 0
34 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
38 42 1 0 0 0 0
39 43 1 0 0 0 0
40 44 1 0 0 0 0
S SKP 8
ID FL63AGNS0013
KNApSAcK_ID C00008891
NAME Gallocatechin 7,3'-di-O-gallate
CAS_RN 142784-35-8
FORMULA C29H22O15
EXACTMASS 610.095870034
AVERAGEMASS 610.4759799999999
SMILES C(c(c5)cc(c(O)c5O)O)(Oc(c4)cc(c(c(O)4)3)OC(C(C3)O)c(c2)cc(c(O)c2O)OC(c(c1)cc(c(c1O)O)O)=O)=O
M END