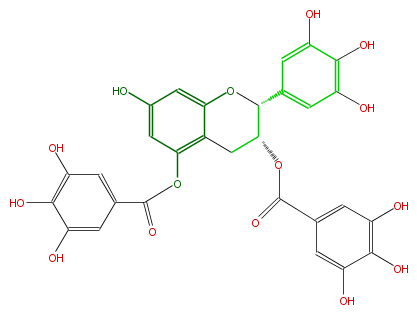
Mol:FL63AGNS0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1792 1.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1792 1.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1792 0.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1792 0.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6407 0.0879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6407 0.0879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1022 0.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1022 0.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1022 1.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1022 1.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6407 1.3316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6407 1.3316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4363 0.0879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4363 0.0879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9749 0.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9749 0.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9749 1.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9749 1.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4363 1.3316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4363 1.3316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7175 1.3315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7175 1.3315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4544 1.2976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4544 1.2976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9991 0.9831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9991 0.9831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5438 1.2976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5438 1.2976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5438 1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5438 1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9991 2.2410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9991 2.2410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4544 1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4544 1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6407 -0.5336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6407 -0.5336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9991 2.8694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9991 2.8694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3968 -0.1510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3968 -0.1510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3989 -0.8355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3989 -0.8355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9320 -1.3025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9320 -1.3025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1365 -1.9248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1365 -1.9248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6819 -2.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6819 -2.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2273 -1.9248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2273 -1.9248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2273 -1.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2273 -1.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6819 -0.9800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6819 -0.9800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1365 -1.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1365 -1.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7727 -0.9801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7727 -0.9801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7727 -2.2396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7727 -2.2396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6819 -2.8694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6819 -2.8694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0880 2.2407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0880 2.2407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0880 0.9834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0880 0.9834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2951 -0.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2951 -0.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1410 -1.4866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1410 -1.4866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8824 -0.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8824 -0.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1976 -0.3656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1976 -0.3656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8278 -0.3656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8278 -0.3656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1430 -0.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1430 -0.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8278 -1.4572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8278 -1.4572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1976 -1.4572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1976 -1.4572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1427 0.1798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1427 0.1798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7727 -0.9114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7727 -0.9114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1427 -2.0026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1427 -2.0026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 1 11 1 0 0 0 0 | + | 1 11 1 0 0 0 0 |
| − | 9 12 1 6 0 0 0 | + | 9 12 1 6 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 2 0 0 0 0 | + | 21 22 2 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 25 30 1 0 0 0 0 | + | 25 30 1 0 0 0 0 |
| − | 28 21 1 0 0 0 0 | + | 28 21 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 8 20 1 6 0 0 0 | + | 8 20 1 6 0 0 0 |
| − | 15 32 1 0 0 0 0 | + | 15 32 1 0 0 0 0 |
| − | 14 33 1 0 0 0 0 | + | 14 33 1 0 0 0 0 |
| − | 18 34 1 0 0 0 0 | + | 18 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 40 44 1 0 0 0 0 | + | 40 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL63AGNS0009 | + | ID FL63AGNS0009 |
| − | KNApSAcK_ID C00008887 | + | KNApSAcK_ID C00008887 |
| − | NAME Epigallocatechin 3,5,-di-O-gallate | + | NAME Epigallocatechin 3,5,-di-O-gallate |
| − | CAS_RN 37484-73-4 | + | CAS_RN 37484-73-4 |
| − | FORMULA C29H22O15 | + | FORMULA C29H22O15 |
| − | EXACTMASS 610.095870034 | + | EXACTMASS 610.095870034 |
| − | AVERAGEMASS 610.4759799999999 | + | AVERAGEMASS 610.4759799999999 |
| − | SMILES C(c32)C(C(c(c5)cc(c(c5O)O)O)Oc(cc(cc3OC(c(c4)cc(c(c4O)O)O)=O)O)2)OC(c(c1)cc(c(c1O)O)O)=O | + | SMILES C(c32)C(C(c(c5)cc(c(c5O)O)O)Oc(cc(cc3OC(c(c4)cc(c(c4O)O)O)=O)O)2)OC(c(c1)cc(c(c1O)O)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-1.1792 1.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1792 0.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6407 0.0879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1022 0.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1022 1.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6407 1.3316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4363 0.0879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9749 0.3989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9749 1.0207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4363 1.3316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7175 1.3315 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4544 1.2976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9991 0.9831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5438 1.2976 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5438 1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9991 2.2410 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4544 1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6407 -0.5336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9991 2.8694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3968 -0.1510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3989 -0.8355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9320 -1.3025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1365 -1.9248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6819 -2.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2273 -1.9248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2273 -1.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6819 -0.9800 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1365 -1.2950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7727 -0.9801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7727 -2.2396 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6819 -2.8694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0880 2.2407 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0880 0.9834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2951 -0.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1410 -1.4866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8824 -0.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1976 -0.3656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8278 -0.3656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1430 -0.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8278 -1.4572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1976 -1.4572 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1427 0.1798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7727 -0.9114 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1427 -2.0026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
1 11 1 0 0 0 0
9 12 1 6 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 0 0 0 0
21 22 2 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
26 29 1 0 0 0 0
25 30 1 0 0 0 0
28 21 1 0 0 0 0
24 31 1 0 0 0 0
8 20 1 6 0 0 0
15 32 1 0 0 0 0
14 33 1 0 0 0 0
18 34 1 0 0 0 0
34 35 2 0 0 0 0
34 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
38 42 1 0 0 0 0
39 43 1 0 0 0 0
40 44 1 0 0 0 0
S SKP 8
ID FL63AGNS0009
KNApSAcK_ID C00008887
NAME Epigallocatechin 3,5,-di-O-gallate
CAS_RN 37484-73-4
FORMULA C29H22O15
EXACTMASS 610.095870034
AVERAGEMASS 610.4759799999999
SMILES C(c32)C(C(c(c5)cc(c(c5O)O)O)Oc(cc(cc3OC(c(c4)cc(c(c4O)O)O)=O)O)2)OC(c(c1)cc(c(c1O)O)O)=O
M END