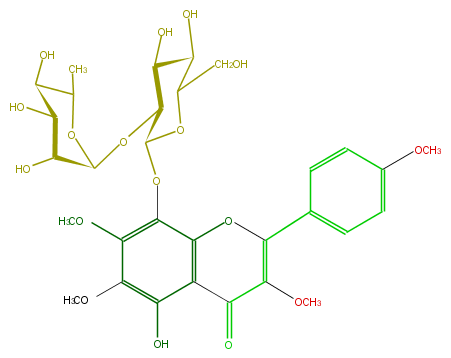
Mol:FL5FGAGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 48 52 0 0 0 0 0 0 0 0999 V2000 | + | 48 52 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.4094 -1.0863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4094 -1.0863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4094 -1.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4094 -1.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6951 -2.3240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6951 -2.3240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9805 -1.9117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9805 -1.9117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9805 -1.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9805 -1.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6951 -0.6739 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6951 -0.6739 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2661 -2.3240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2661 -2.3240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4484 -1.9116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4484 -1.9116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4485 -1.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4485 -1.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2660 -0.6739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2660 -0.6739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2661 -3.1606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2661 -3.1606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3482 -0.5152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3482 -0.5152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0764 -0.9356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0764 -0.9356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8046 -0.5152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8046 -0.5152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8046 0.3257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8046 0.3257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0765 0.7461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0765 0.7461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3482 0.3258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3482 0.3258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6951 -3.1488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6951 -3.1488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9754 2.5771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9754 2.5771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7616 2.3991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7616 2.3991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6252 1.5633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6252 1.5633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9452 0.8769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9452 0.8769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2335 1.1359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2335 1.1359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2954 1.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2954 1.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1078 3.4594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1078 3.4594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6098 3.1091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6098 3.1091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3762 0.9094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3762 0.9094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7104 0.1210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7104 0.1210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7571 0.5784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7571 0.5784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9622 0.3548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9622 0.3548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4055 1.0449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4055 1.0449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3939 1.8280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3939 1.8280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1462 2.0296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1462 2.0296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7457 1.3616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7457 1.3616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9746 2.6412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9746 2.6412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4131 2.3523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4131 2.3523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4341 1.6077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4341 1.6077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3079 0.3645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3079 0.3645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5166 0.7368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5166 0.7368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4341 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4341 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4743 2.4626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4743 2.4626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5778 3.1992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5778 3.1992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1222 -0.6747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1222 -0.6747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3869 -0.4518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3869 -0.4518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0235 -2.2660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0235 -2.2660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9411 -2.7959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9411 -2.7959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1182 -2.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1182 -2.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6673 -3.4594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6673 -3.4594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 28 6 1 0 0 0 0 | + | 28 6 1 0 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 29 1 1 0 0 0 | + | 34 29 1 1 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 34 37 1 0 0 0 0 | + | 34 37 1 0 0 0 0 |
| − | 29 38 1 0 0 0 0 | + | 29 38 1 0 0 0 0 |
| − | 30 27 1 0 0 0 0 | + | 30 27 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 15 39 1 0 0 0 0 | + | 15 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 24 41 1 0 0 0 0 | + | 24 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 1 43 1 0 0 0 0 | + | 1 43 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 2 45 1 0 0 0 0 | + | 2 45 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 8 47 1 0 0 0 0 | + | 8 47 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 44 -0.7120 -0.4111 | + | M SBV 1 44 -0.7120 -0.4111 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 46 -0.8211 -0.5719 | + | M SBV 2 46 -0.8211 -0.5719 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 ^ OCH3 | + | M SMT 3 ^ OCH3 |
| − | M SBV 3 48 0.7128 -0.4115 | + | M SBV 3 48 0.7128 -0.4115 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 45 46 | + | M SAL 4 2 45 46 |
| − | M SBL 4 1 50 | + | M SBL 4 1 50 |
| − | M SMT 4 ^ OCH3 | + | M SMT 4 ^ OCH3 |
| − | M SBV 4 50 0.6142 0.3546 | + | M SBV 4 50 0.6142 0.3546 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 47 48 | + | M SAL 5 2 47 48 |
| − | M SBL 5 1 52 | + | M SBL 5 1 52 |
| − | M SMT 5 OCH3 | + | M SMT 5 OCH3 |
| − | M SBV 5 52 -0.6698 0.3867 | + | M SBV 5 52 -0.6698 0.3867 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FGAGS0001 | + | ID FL5FGAGS0001 |
| − | FORMULA C31H38O17 | + | FORMULA C31H38O17 |
| − | EXACTMASS 682.21089979 | + | EXACTMASS 682.21089979 |
| − | AVERAGEMASS 682.62322 | + | AVERAGEMASS 682.62322 |
| − | SMILES O(C(C5O)OC(C(O)C(O)5)C)C(C1Oc(c24)c(c(c(O)c2C(C(=C(O4)c(c3)ccc(c3)OC)OC)=O)OC)OC)C(C(O)C(O1)CO)O | + | SMILES O(C(C5O)OC(C(O)C(O)5)C)C(C1Oc(c24)c(c(c(O)c2C(C(=C(O4)c(c3)ccc(c3)OC)OC)=O)OC)OC)C(C(O)C(O1)CO)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
48 52 0 0 0 0 0 0 0 0999 V2000
-2.4094 -1.0863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4094 -1.9114 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6951 -2.3240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9805 -1.9117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9805 -1.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6951 -0.6739 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2661 -2.3240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4484 -1.9116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4485 -1.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2660 -0.6739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2661 -3.1606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3482 -0.5152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0764 -0.9356 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8046 -0.5152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8046 0.3257 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0765 0.7461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3482 0.3258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6951 -3.1488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9754 2.5771 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7616 2.3991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6252 1.5633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9452 0.8769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2335 1.1359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2954 1.8908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1078 3.4594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6098 3.1091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3762 0.9094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7104 0.1210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7571 0.5784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9622 0.3548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4055 1.0449 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3939 1.8280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1462 2.0296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7457 1.3616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9746 2.6412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4131 2.3523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4341 1.6077 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3079 0.3645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5166 0.7368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4341 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4743 2.4626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5778 3.1992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1222 -0.6747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3869 -0.4518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0235 -2.2660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9411 -2.7959 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1182 -2.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6673 -3.4594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
4 3 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
28 6 1 0 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 29 1 1 0 0 0
33 35 1 0 0 0 0
32 36 1 0 0 0 0
34 37 1 0 0 0 0
29 38 1 0 0 0 0
30 27 1 0 0 0 0
39 40 1 0 0 0 0
15 39 1 0 0 0 0
41 42 1 0 0 0 0
24 41 1 0 0 0 0
43 44 1 0 0 0 0
1 43 1 0 0 0 0
45 46 1 0 0 0 0
2 45 1 0 0 0 0
47 48 1 0 0 0 0
8 47 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 OCH3
M SBV 1 44 -0.7120 -0.4111
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 CH2OH
M SBV 2 46 -0.8211 -0.5719
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 48
M SMT 3 ^ OCH3
M SBV 3 48 0.7128 -0.4115
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 45 46
M SBL 4 1 50
M SMT 4 ^ OCH3
M SBV 4 50 0.6142 0.3546
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 47 48
M SBL 5 1 52
M SMT 5 OCH3
M SBV 5 52 -0.6698 0.3867
S SKP 5
ID FL5FGAGS0001
FORMULA C31H38O17
EXACTMASS 682.21089979
AVERAGEMASS 682.62322
SMILES O(C(C5O)OC(C(O)C(O)5)C)C(C1Oc(c24)c(c(c(O)c2C(C(=C(O4)c(c3)ccc(c3)OC)OC)=O)OC)OC)C(C(O)C(O1)CO)O
M END