Mol:FL5FFCGA0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 36 39 0 0 0 0 0 0 0 0999 V2000 | + | 36 39 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2472 -4.3461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2472 -4.3461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6268 -4.5124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6268 -4.5124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1726 -4.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1726 -4.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3389 -3.4377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3389 -3.4377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9593 -3.2714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9593 -3.2714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4135 -3.7256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4135 -3.7256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1154 -2.9835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1154 -2.9835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0509 -2.3630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0509 -2.3630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6713 -2.1967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6713 -2.1967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1256 -2.6510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1256 -2.6510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5991 -3.1130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5991 -3.1130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8376 -1.5765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8376 -1.5765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3746 -1.1135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3746 -1.1135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5441 -0.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5441 -0.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1764 -0.3117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1764 -0.3117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6393 -0.7746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6393 -0.7746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4699 -1.4070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4699 -1.4070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7013 -4.8002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7013 -4.8002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6484 -1.8794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6484 -1.8794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3802 -0.6192 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.3802 -0.6192 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.9925 -1.2906 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9925 -1.2906 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.7380 -1.0776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7380 -1.0776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4879 -1.2906 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.4879 -1.2906 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.8757 -0.6192 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.8757 -0.6192 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1302 -0.8322 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.1302 -0.8322 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.6531 -0.8140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6531 -0.8140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3990 -0.3666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3990 -0.3666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6344 0.2811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6344 0.2811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4353 0.6542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4353 0.6542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4476 -4.2242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4476 -4.2242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8950 -3.3087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8950 -3.3087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6510 -2.6541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6510 -2.6541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1225 -0.3518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1225 -0.3518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8749 0.3068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8749 0.3068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0947 -1.9612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0947 -1.9612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2978 -1.7477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2978 -1.7477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 21 19 1 0 0 0 0 | + | 21 19 1 0 0 0 0 |
| − | 19 8 1 0 0 0 0 | + | 19 8 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 15 29 1 0 0 0 0 | + | 15 29 1 0 0 0 0 |
| − | 3 30 1 0 0 0 0 | + | 3 30 1 0 0 0 0 |
| − | 6 31 1 0 0 0 0 | + | 6 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 16 33 1 0 0 0 0 | + | 16 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 23 35 1 0 0 0 0 | + | 23 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 35 36 | + | M SAL 3 2 35 36 |
| − | M SBL 3 1 38 | + | M SBL 3 1 38 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 38 3.0947 -1.9612 | + | M SVB 3 38 3.0947 -1.9612 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 33 34 | + | M SAL 2 2 33 34 |
| − | M SBL 2 1 36 | + | M SBL 2 1 36 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 36 1.0931 1.9612 | + | M SVB 2 36 1.0931 1.9612 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 31 32 | + | M SAL 1 2 31 32 |
| − | M SBL 1 1 34 | + | M SBL 1 1 34 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 34 -1.7042 0.9762 | + | M SVB 1 34 -1.7042 0.9762 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FFCGA0006 | + | ID FL5FFCGA0006 |
| − | KNApSAcK_ID C00005714 | + | KNApSAcK_ID C00005714 |
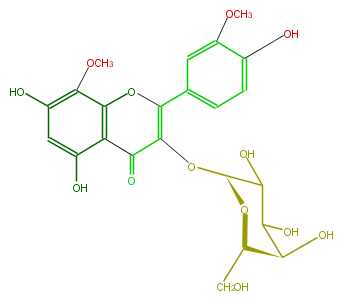
| − | NAME Limocitrin 3-galactoside | + | NAME Limocitrin 3-galactoside |
| − | CAS_RN 103839-19-6 | + | CAS_RN 103839-19-6 |
| − | FORMULA C23H24O13 | + | FORMULA C23H24O13 |
| − | EXACTMASS 508.121690854 | + | EXACTMASS 508.121690854 |
| − | AVERAGEMASS 508.42886 | + | AVERAGEMASS 508.42886 |
| − | SMILES O(c(c(O)4)cc(cc4)C(=C(O[C@H](O3)C(O)C([C@H]([C@@H](CO)3)O)O)2)Oc(c1C2=O)c(OC)c(cc(O)1)O)C | + | SMILES O(c(c(O)4)cc(cc4)C(=C(O[C@H](O3)C(O)C([C@H]([C@@H](CO)3)O)O)2)Oc(c1C2=O)c(OC)c(cc(O)1)O)C |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
36 39 0 0 0 0 0 0 0 0999 V2000
-1.2472 -4.3461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6268 -4.5124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1726 -4.0582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3389 -3.4377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9593 -3.2714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4135 -3.7256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1154 -2.9835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0509 -2.3630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6713 -2.1967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1256 -2.6510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5991 -3.1130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8376 -1.5765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3746 -1.1135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5441 -0.4811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1764 -0.3117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6393 -0.7746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4699 -1.4070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7013 -4.8002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6484 -1.8794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3802 -0.6192 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.9925 -1.2906 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.7380 -1.0776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4879 -1.2906 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8757 -0.6192 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1302 -0.8322 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.6531 -0.8140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3990 -0.3666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6344 0.2811 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4353 0.6542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4476 -4.2242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8950 -3.3087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6510 -2.6541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1225 -0.3518 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8749 0.3068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0947 -1.9612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2978 -1.7477 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
21 19 1 0 0 0 0
19 8 1 0 0 0 0
23 22 1 1 0 0 0
15 29 1 0 0 0 0
3 30 1 0 0 0 0
6 31 1 0 0 0 0
31 32 1 0 0 0 0
16 33 1 0 0 0 0
33 34 1 0 0 0 0
23 35 1 0 0 0 0
35 36 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 35 36
M SBL 3 1 38
M SMT 3 CH2OH
M SVB 3 38 3.0947 -1.9612
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 33 34
M SBL 2 1 36
M SMT 2 OCH3
M SVB 2 36 1.0931 1.9612
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 31 32
M SBL 1 1 34
M SMT 1 OCH3
M SVB 1 34 -1.7042 0.9762
S SKP 8
ID FL5FFCGA0006
KNApSAcK_ID C00005714
NAME Limocitrin 3-galactoside
CAS_RN 103839-19-6
FORMULA C23H24O13
EXACTMASS 508.121690854
AVERAGEMASS 508.42886
SMILES O(c(c(O)4)cc(cc4)C(=C(O[C@H](O3)C(O)C([C@H]([C@@H](CO)3)O)O)2)Oc(c1C2=O)c(OC)c(cc(O)1)O)C
M END