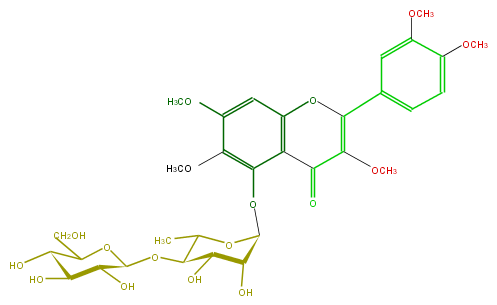
Mol:FL5FECGS0037
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 49 53 0 0 0 0 0 0 0 0999 V2000 | + | 49 53 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1141 0.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1141 0.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1141 -0.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1141 -0.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3996 -0.9670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3996 -0.9670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3150 -0.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3150 -0.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3150 0.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3150 0.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3996 0.6831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3996 0.6831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0295 -0.9670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0295 -0.9670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7440 -0.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7440 -0.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7440 0.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7440 0.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0295 0.6831 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0295 0.6831 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0295 -1.7783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0295 -1.7783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6438 0.8419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6438 0.8419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3721 0.4215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3721 0.4215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1004 0.8419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1004 0.8419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1004 1.6828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1004 1.6828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3721 2.1033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3721 2.1033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6438 1.6828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6438 1.6828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3996 -1.7919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3996 -1.7919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2269 -2.9500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2269 -2.9500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7501 -3.5792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7501 -3.5792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0636 -3.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0636 -3.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4012 -3.3051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4012 -3.3051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8826 -2.8236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8826 -2.8236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4832 -3.1406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4832 -3.1406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9433 -3.2561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9433 -3.2561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4669 -3.5750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4669 -3.5750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4599 -3.7499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4599 -3.7499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6911 -2.5620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6911 -2.5620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0652 -3.2098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0652 -3.2098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3459 -3.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3459 -3.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6223 -3.2098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6223 -3.2098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2483 -2.5620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2483 -2.5620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9675 -2.7675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9675 -2.7675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8723 -3.5753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8723 -3.5753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7284 -3.0461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7284 -3.0461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4019 -2.6519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4019 -2.6519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6771 -3.8918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6771 -3.8918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3735 2.7393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3735 2.7393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9404 3.8918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9404 3.8918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7875 0.6593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7875 0.6593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4296 1.7718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4296 1.7718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4966 -0.9889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4966 -0.9889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0458 -2.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0458 -2.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1842 -2.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1842 -2.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5467 -1.5336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5467 -1.5336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6588 2.0052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6588 2.0052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9433 2.0052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9433 2.0052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8039 -0.9527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8039 -0.9527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7996 -0.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7996 -0.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 19 20 1 1 0 0 0 | + | 19 20 1 1 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 30 34 1 0 0 0 0 | + | 30 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 28 36 1 0 0 0 0 | + | 28 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 18 32 1 0 0 0 0 | + | 18 32 1 0 0 0 0 |
| − | 35 22 1 0 0 0 0 | + | 35 22 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 16 38 1 0 0 0 0 | + | 16 38 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 1 40 1 0 0 0 0 | + | 1 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 8 42 1 0 0 0 0 | + | 8 42 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 24 44 1 0 0 0 0 | + | 24 44 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 15 46 1 0 0 0 0 | + | 15 46 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 2 48 1 0 0 0 0 | + | 2 48 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 43 -0.0014 -0.6359 | + | M SBV 1 43 -0.0014 -0.6359 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 ^ OCH3 | + | M SMT 2 ^ OCH3 |
| − | M SBV 2 45 0.6734 -0.3888 | + | M SBV 2 45 0.6734 -0.3888 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 42 43 | + | M SAL 3 2 42 43 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 47 -0.7526 0.4345 | + | M SBV 3 47 -0.7526 0.4345 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 44 45 | + | M SAL 4 2 44 45 |
| − | M SBL 4 1 49 | + | M SBL 4 1 49 |
| − | M SMT 4 ^ CH2OH | + | M SMT 4 ^ CH2OH |
| − | M SBV 4 49 0.7010 -0.6113 | + | M SBV 4 49 0.7010 -0.6113 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 46 47 | + | M SAL 5 2 46 47 |
| − | M SBL 5 1 51 | + | M SBL 5 1 51 |
| − | M SMT 5 OCH3 | + | M SMT 5 OCH3 |
| − | M SBV 5 51 -0.5585 -0.3224 | + | M SBV 5 51 -0.5585 -0.3224 |
| − | M STY 1 6 SUP | + | M STY 1 6 SUP |
| − | M SLB 1 6 6 | + | M SLB 1 6 6 |
| − | M SAL 6 2 48 49 | + | M SAL 6 2 48 49 |
| − | M SBL 6 1 53 | + | M SBL 6 1 53 |
| − | M SMT 6 ^ OCH3 | + | M SMT 6 ^ OCH3 |
| − | M SBV 6 53 0.6898 0.3983 | + | M SBV 6 53 0.6898 0.3983 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FECGS0037 | + | ID FL5FECGS0037 |
| − | FORMULA C32H40O17 | + | FORMULA C32H40O17 |
| − | EXACTMASS 696.226549854 | + | EXACTMASS 696.226549854 |
| − | AVERAGEMASS 696.6497999999999 | + | AVERAGEMASS 696.6497999999999 |
| − | SMILES O(C(OC(C5C)C(C(C(O5)Oc(c(OC)4)c(C2=O)c(cc(OC)4)OC(c(c3)cc(c(OC)c3)OC)=C2OC)O)O)1)C(C(O)C(O)C1O)CO | + | SMILES O(C(OC(C5C)C(C(C(O5)Oc(c(OC)4)c(C2=O)c(cc(OC)4)OC(c(c3)cc(c(OC)c3)OC)=C2OC)O)O)1)C(C(O)C(O)C1O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
49 53 0 0 0 0 0 0 0 0999 V2000
-1.1141 0.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1141 -0.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3996 -0.9670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3150 -0.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3150 0.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3996 0.6831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0295 -0.9670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7440 -0.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7440 0.2706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0295 0.6831 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0295 -1.7783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6438 0.8419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3721 0.4215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1004 0.8419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1004 1.6828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3721 2.1033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6438 1.6828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3996 -1.7919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2269 -2.9500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7501 -3.5792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0636 -3.3123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4012 -3.3051 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8826 -2.8236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4832 -3.1406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9433 -3.2561 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4669 -3.5750 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4599 -3.7499 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6911 -2.5620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0652 -3.2098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3459 -3.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6223 -3.2098 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2483 -2.5620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9675 -2.7675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8723 -3.5753 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7284 -3.0461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4019 -2.6519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6771 -3.8918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3735 2.7393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9404 3.8918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7875 0.6593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4296 1.7718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4966 -0.9889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0458 -2.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1842 -2.5293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5467 -1.5336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6588 2.0052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9433 2.0052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8039 -0.9527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7996 -0.5905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
4 3 1 0 0 0 0
19 20 1 1 0 0 0
20 21 1 1 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
30 34 1 0 0 0 0
29 35 1 0 0 0 0
28 36 1 0 0 0 0
31 37 1 0 0 0 0
18 32 1 0 0 0 0
35 22 1 0 0 0 0
38 39 1 0 0 0 0
16 38 1 0 0 0 0
40 41 1 0 0 0 0
1 40 1 0 0 0 0
42 43 1 0 0 0 0
8 42 1 0 0 0 0
44 45 1 0 0 0 0
24 44 1 0 0 0 0
46 47 1 0 0 0 0
15 46 1 0 0 0 0
48 49 1 0 0 0 0
2 48 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 43
M SMT 1 OCH3
M SBV 1 43 -0.0014 -0.6359
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 45
M SMT 2 ^ OCH3
M SBV 2 45 0.6734 -0.3888
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 42 43
M SBL 3 1 47
M SMT 3 OCH3
M SBV 3 47 -0.7526 0.4345
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 44 45
M SBL 4 1 49
M SMT 4 ^ CH2OH
M SBV 4 49 0.7010 -0.6113
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 46 47
M SBL 5 1 51
M SMT 5 OCH3
M SBV 5 51 -0.5585 -0.3224
M STY 1 6 SUP
M SLB 1 6 6
M SAL 6 2 48 49
M SBL 6 1 53
M SMT 6 ^ OCH3
M SBV 6 53 0.6898 0.3983
S SKP 5
ID FL5FECGS0037
FORMULA C32H40O17
EXACTMASS 696.226549854
AVERAGEMASS 696.6497999999999
SMILES O(C(OC(C5C)C(C(C(O5)Oc(c(OC)4)c(C2=O)c(cc(OC)4)OC(c(c3)cc(c(OC)c3)OC)=C2OC)O)O)1)C(C(O)C(O)C1O)CO
M END