Mol:FL5FECGS0036
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.9579 -1.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9579 -1.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9579 -1.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9579 -1.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4016 -2.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4016 -2.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8453 -1.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8453 -1.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8453 -1.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8453 -1.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4016 -0.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4016 -0.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2890 -2.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2890 -2.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2673 -1.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2673 -1.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2673 -1.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2673 -1.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2890 -0.9126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2890 -0.9126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2890 -2.6982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2890 -2.6982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8234 -0.9127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8234 -0.9127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3904 -1.2401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3904 -1.2401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9574 -0.9127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9574 -0.9127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9574 -0.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9574 -0.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3904 0.0693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3904 0.0693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8234 -0.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8234 -0.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4016 -2.8394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4016 -2.8394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3904 0.7238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3904 0.7238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2405 2.5098 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 0.2405 2.5098 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -0.0248 1.8803 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.0248 1.8803 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.4723 1.4813 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.4723 1.4813 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.7642 0.9881 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.7642 0.9881 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.9167 1.5571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9167 1.5571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4194 1.8700 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.4194 1.8700 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.2404 3.1691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2404 3.1691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6635 1.9611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6635 1.9611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1476 0.9009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1476 0.9009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5415 0.0609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5415 0.0609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2560 -0.3516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2560 -0.3516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2583 1.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2583 1.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7328 2.7748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7328 2.7748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5415 -2.4167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5415 -2.4167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2560 -2.0042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2560 -2.0042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1333 -2.3761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1333 -2.3761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9994 -2.8760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9994 -2.8760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3151 -0.6150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3151 -0.6150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8150 0.2511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8150 0.2511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 19 23 1 0 0 0 0 | + | 19 23 1 0 0 0 0 |
| − | 15 29 1 0 0 0 0 | + | 15 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 2 33 1 0 0 0 0 | + | 2 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 8 35 1 0 0 0 0 | + | 8 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 1 37 1 0 0 0 0 | + | 1 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 31 32 | + | M SAL 5 2 31 32 |
| − | M SBL 5 1 34 | + | M SBL 5 1 34 |
| − | M SMT 5 CH2OH | + | M SMT 5 CH2OH |
| − | M SVB 5 34 1.2583 1.8945 | + | M SVB 5 34 1.2583 1.8945 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 37 38 | + | M SAL 4 2 37 38 |
| − | M SBL 4 1 40 | + | M SBL 4 1 40 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SVB 4 40 -2.3151 -0.615 | + | M SVB 4 40 -2.3151 -0.615 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 35 36 | + | M SAL 3 2 35 36 |
| − | M SBL 3 1 38 | + | M SBL 3 1 38 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 38 0.4662 -2.0824 | + | M SVB 3 38 0.4662 -2.0824 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 33 34 | + | M SAL 2 2 33 34 |
| − | M SBL 2 1 36 | + | M SBL 2 1 36 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 36 -2.5415 -2.4167 | + | M SVB 2 36 -2.5415 -2.4167 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 29 30 | + | M SAL 1 2 29 30 |
| − | M SBL 1 1 32 | + | M SBL 1 1 32 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 32 2.5415 0.0609 | + | M SVB 1 32 2.5415 0.0609 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FECGS0036 | + | ID FL5FECGS0036 |
| − | KNApSAcK_ID C00005683 | + | KNApSAcK_ID C00005683 |
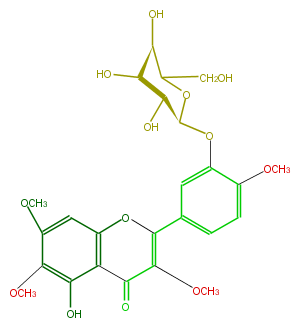
| − | NAME Casticin 3'-glucoside | + | NAME Casticin 3'-glucoside |
| − | CAS_RN 71827-14-0 | + | CAS_RN 71827-14-0 |
| − | FORMULA C25H28O13 | + | FORMULA C25H28O13 |
| − | EXACTMASS 536.152990982 | + | EXACTMASS 536.152990982 |
| − | AVERAGEMASS 536.48202 | + | AVERAGEMASS 536.48202 |
| − | SMILES O(C)c(c3O[C@H](O4)[C@H]([C@@H](O)[C@H](C4CO)O)O)ccc(c3)C(=C2OC)Oc(c1)c(C2=O)c(O)c(OC)c1OC | + | SMILES O(C)c(c3O[C@H](O4)[C@H]([C@@H](O)[C@H](C4CO)O)O)ccc(c3)C(=C2OC)Oc(c1)c(C2=O)c(O)c(OC)c1OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-1.9579 -1.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9579 -1.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4016 -2.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8453 -1.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8453 -1.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4016 -0.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2890 -2.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2673 -1.8761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2673 -1.2338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2890 -0.9126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2890 -2.6982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8234 -0.9127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3904 -1.2401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9574 -0.9127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9574 -0.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3904 0.0693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8234 -0.2580 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4016 -2.8394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3904 0.7238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2405 2.5098 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.0248 1.8803 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.4723 1.4813 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.7642 0.9881 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.9167 1.5571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4194 1.8700 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.2404 3.1691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6635 1.9611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1476 0.9009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5415 0.0609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2560 -0.3516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2583 1.8945 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7328 2.7748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5415 -2.4167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2560 -2.0042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1333 -2.3761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9994 -2.8760 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3151 -0.6150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8150 0.2511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
19 23 1 0 0 0 0
15 29 1 0 0 0 0
29 30 1 0 0 0 0
25 31 1 0 0 0 0
31 32 1 0 0 0 0
2 33 1 0 0 0 0
33 34 1 0 0 0 0
8 35 1 0 0 0 0
35 36 1 0 0 0 0
1 37 1 0 0 0 0
37 38 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 31 32
M SBL 5 1 34
M SMT 5 CH2OH
M SVB 5 34 1.2583 1.8945
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 37 38
M SBL 4 1 40
M SMT 4 OCH3
M SVB 4 40 -2.3151 -0.615
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 35 36
M SBL 3 1 38
M SMT 3 OCH3
M SVB 3 38 0.4662 -2.0824
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 33 34
M SBL 2 1 36
M SMT 2 OCH3
M SVB 2 36 -2.5415 -2.4167
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 29 30
M SBL 1 1 32
M SMT 1 OCH3
M SVB 1 32 2.5415 0.0609
S SKP 8
ID FL5FECGS0036
KNApSAcK_ID C00005683
NAME Casticin 3'-glucoside
CAS_RN 71827-14-0
FORMULA C25H28O13
EXACTMASS 536.152990982
AVERAGEMASS 536.48202
SMILES O(C)c(c3O[C@H](O4)[C@H]([C@@H](O)[C@H](C4CO)O)O)ccc(c3)C(=C2OC)Oc(c1)c(C2=O)c(O)c(OC)c1OC
M END