Mol:FL5FECGS0031
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 48 52 0 0 0 0 0 0 0 0999 V2000 | + | 48 52 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.9181 -1.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9181 -1.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9181 -1.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9181 -1.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3618 -2.0824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3618 -2.0824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1945 -1.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1945 -1.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1945 -1.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1945 -1.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3618 -0.7977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3618 -0.7977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7508 -2.0824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7508 -2.0824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3071 -1.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3071 -1.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3071 -1.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3071 -1.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7508 -0.7977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7508 -0.7977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7508 -3.0824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7508 -3.0824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0076 -0.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0076 -0.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5746 -1.0014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5746 -1.0014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1416 -0.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1416 -0.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1416 -0.0194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1416 -0.0194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5746 0.3080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5746 0.3080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0076 -0.0194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0076 -0.0194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3618 -2.7245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3618 -2.7245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7484 0.3310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7484 0.3310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4742 -2.0823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4742 -2.0823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0891 1.4252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0891 1.4252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6570 1.0973 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.6570 1.0973 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.4768 1.7278 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.4768 1.7278 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.6570 2.3622 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.6570 2.3622 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.0891 2.6901 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.0891 2.6901 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2692 2.0595 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 3.2692 2.0595 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 3.0444 3.2004 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0444 3.2004 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0114 2.5668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0114 2.5668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8944 1.4867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8944 1.4867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5518 -2.2562 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.5518 -2.2562 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.1806 -2.7462 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.1806 -2.7462 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.6461 -2.5383 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.6461 -2.5383 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1304 -2.5327 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1304 -2.5327 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5051 -2.1579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5051 -2.1579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9727 -2.4047 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.9727 -2.4047 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.1427 -2.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1427 -2.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5310 -3.2074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5310 -3.2074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3398 -3.0524 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3398 -3.0524 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2753 -0.5001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2753 -0.5001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7752 0.3660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7752 0.3660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0077 -1.1739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0077 -1.1739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8737 -1.6739 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8737 -1.6739 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7056 -2.6036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7056 -2.6036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1332 -3.5076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1332 -3.5076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5930 2.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5930 2.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8511 1.6072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8511 1.6072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6653 -1.8624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6653 -1.8624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9475 -1.0871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9475 -1.0871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 2 20 1 0 0 0 0 | + | 2 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 19 22 1 0 0 0 0 | + | 19 22 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 1 39 1 0 0 0 0 | + | 1 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 14 41 1 0 0 0 0 | + | 14 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 8 43 1 0 0 0 0 | + | 8 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 26 45 1 0 0 0 0 | + | 26 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 35 47 1 0 0 0 0 | + | 35 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 47 48 | + | M SAL 5 2 47 48 |
| − | M SBL 5 1 51 | + | M SBL 5 1 51 |
| − | M SMT 5 CH2OH | + | M SMT 5 CH2OH |
| − | M SVB 5 51 -3.3712 -1.6205 | + | M SVB 5 51 -3.3712 -1.6205 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 45 46 | + | M SAL 4 2 45 46 |
| − | M SBL 4 1 49 | + | M SBL 4 1 49 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 49 2.7486 2.5431 | + | M SVB 4 49 2.7486 2.5431 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 47 1.7056 -2.6036 | + | M SVB 3 47 1.7056 -2.6036 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 45 3.3511 -0.8803 | + | M SVB 2 45 3.3511 -0.8803 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 43 -1.2753 -0.5001 | + | M SVB 1 43 -1.2753 -0.5001 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FECGS0031 | + | ID FL5FECGS0031 |
| − | KNApSAcK_ID C00005677 | + | KNApSAcK_ID C00005677 |
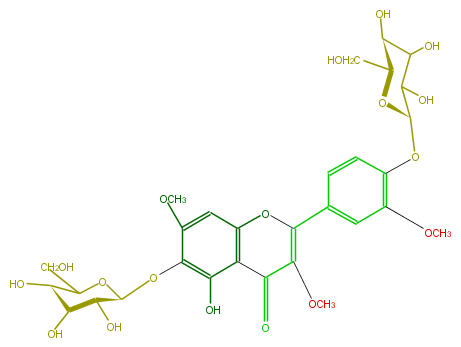
| − | NAME Chrysosplenol C 6,4'-diglucoside | + | NAME Chrysosplenol C 6,4'-diglucoside |
| − | CAS_RN 64190-92-7 | + | CAS_RN 64190-92-7 |
| − | FORMULA C30H36O18 | + | FORMULA C30H36O18 |
| − | EXACTMASS 684.190164348 | + | EXACTMASS 684.190164348 |
| − | AVERAGEMASS 684.59604 | + | AVERAGEMASS 684.59604 |
| − | SMILES C([C@H](O1)[C@H](C(C([C@H]1Oc(c(OC)2)ccc(C(=C(OC)5)Oc(c4C5=O)cc(c(c(O)4)O[C@@H]([C@H]3O)OC(CO)[C@@H]([C@@H]3O)O)OC)c2)O)O)O)O | + | SMILES C([C@H](O1)[C@H](C(C([C@H]1Oc(c(OC)2)ccc(C(=C(OC)5)Oc(c4C5=O)cc(c(c(O)4)O[C@@H]([C@H]3O)OC(CO)[C@@H]([C@@H]3O)O)OC)c2)O)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
48 52 0 0 0 0 0 0 0 0999 V2000
-0.9181 -1.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9181 -1.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3618 -2.0824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1945 -1.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1945 -1.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3618 -0.7977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7508 -2.0824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3071 -1.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3071 -1.1189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7508 -0.7977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7508 -3.0824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0076 -0.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5746 -1.0014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1416 -0.6740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1416 -0.0194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5746 0.3080 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0076 -0.0194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3618 -2.7245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7484 0.3310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4742 -2.0823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0891 1.4252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6570 1.0973 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.4768 1.7278 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.6570 2.3622 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.0891 2.6901 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2692 2.0595 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
3.0444 3.2004 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0114 2.5668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8944 1.4867 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5518 -2.2562 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.1806 -2.7462 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.6461 -2.5383 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1304 -2.5327 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5051 -2.1579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9727 -2.4047 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.1427 -2.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5310 -3.2074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3398 -3.0524 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2753 -0.5001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7752 0.3660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0077 -1.1739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8737 -1.6739 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7056 -2.6036 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1332 -3.5076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5930 2.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8511 1.6072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6653 -1.8624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9475 -1.0871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
2 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
23 29 1 0 0 0 0
19 22 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 20 1 0 0 0 0
1 39 1 0 0 0 0
39 40 1 0 0 0 0
14 41 1 0 0 0 0
41 42 1 0 0 0 0
8 43 1 0 0 0 0
43 44 1 0 0 0 0
26 45 1 0 0 0 0
45 46 1 0 0 0 0
35 47 1 0 0 0 0
47 48 1 0 0 0 0
M STY 1 5 SUP
M SLB 1 5 5
M SAL 5 2 47 48
M SBL 5 1 51
M SMT 5 CH2OH
M SVB 5 51 -3.3712 -1.6205
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 45 46
M SBL 4 1 49
M SMT 4 CH2OH
M SVB 4 49 2.7486 2.5431
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 47
M SMT 3 OCH3
M SVB 3 47 1.7056 -2.6036
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 OCH3
M SVB 2 45 3.3511 -0.8803
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 OCH3
M SVB 1 43 -1.2753 -0.5001
S SKP 8
ID FL5FECGS0031
KNApSAcK_ID C00005677
NAME Chrysosplenol C 6,4'-diglucoside
CAS_RN 64190-92-7
FORMULA C30H36O18
EXACTMASS 684.190164348
AVERAGEMASS 684.59604
SMILES C([C@H](O1)[C@H](C(C([C@H]1Oc(c(OC)2)ccc(C(=C(OC)5)Oc(c4C5=O)cc(c(c(O)4)O[C@@H]([C@H]3O)OC(CO)[C@@H]([C@@H]3O)O)OC)c2)O)O)O)O
M END