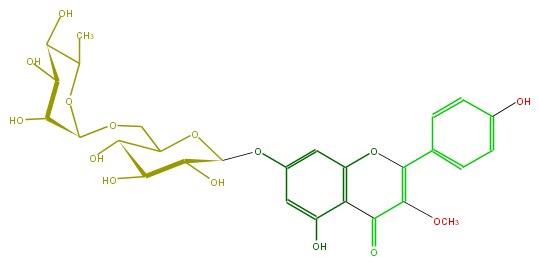
Mol:FL5FDAGS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.4048 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4048 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4048 -1.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4048 -1.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1058 -1.8747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1058 -1.8747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8068 -1.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8068 -1.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8068 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8068 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1058 -0.2558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1058 -0.2558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5078 -1.8747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5078 -1.8747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2088 -1.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2088 -1.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2088 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2088 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5078 -0.2558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5078 -0.2558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5078 -2.6862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5078 -2.6862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9096 -0.2560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9096 -0.2560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6241 -0.6684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6241 -0.6684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3385 -0.2560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3385 -0.2560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3385 0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3385 0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6241 0.9816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6241 0.9816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9096 0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9096 0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2960 -0.2560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2960 -0.2560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1058 -2.5367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1058 -2.5367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0030 0.9526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0030 0.9526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3562 0.5237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3562 0.5237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5645 0.0667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5645 0.0667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8158 0.9456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8158 0.9456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5645 1.8300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5645 1.8300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3562 2.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3562 2.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1051 1.4081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1051 1.4081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9867 3.0720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9867 3.0720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5848 2.5469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5848 2.5469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7704 1.9170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7704 1.9170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0030 0.4875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0030 0.4875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6388 -0.0149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6388 -0.0149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9891 -0.8726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9891 -0.8726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0535 -0.5088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0535 -0.5088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1507 -0.4991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1507 -0.4991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8068 0.1572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8068 0.1572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6253 -0.2749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6253 -0.2749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2468 -0.4132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2468 -0.4132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7738 -0.9440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7738 -0.9440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3511 -0.9976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3511 -0.9976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0885 0.3300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0885 0.3300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7779 0.3628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7779 0.3628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0151 -1.9355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0151 -1.9355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5595 -3.0720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5595 -3.0720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 21 30 1 0 0 0 0 | + | 21 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 18 1 0 0 0 0 | + | 34 18 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 22 41 1 0 0 0 0 | + | 22 41 1 0 0 0 0 |
| − | 41 40 1 0 0 0 0 | + | 41 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 8 42 1 0 0 0 0 | + | 8 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 47 -0.8063 0.4655 | + | M SBV 1 47 -0.8063 0.4655 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FDAGS0003 | + | ID FL5FDAGS0003 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES OC(C(O)1)C(COC(O5)C(C(C(O)C(C)5)O)O)OC(Oc(c2)cc(O)c(C(=O)3)c2OC(c(c4)ccc(O)c4)=C(OC)3)C(O)1 | + | SMILES OC(C(O)1)C(COC(O5)C(C(C(O)C(C)5)O)O)OC(Oc(c2)cc(O)c(C(=O)3)c2OC(c(c4)ccc(O)c4)=C(OC)3)C(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
0.4048 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4048 -1.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1058 -1.8747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8068 -1.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8068 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1058 -0.2558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5078 -1.8747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2088 -1.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2088 -0.6605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5078 -0.2558 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5078 -2.6862 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9096 -0.2560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6241 -0.6684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3385 -0.2560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3385 0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6241 0.9816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9096 0.5690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2960 -0.2560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1058 -2.5367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.0030 0.9526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3562 0.5237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5645 0.0667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8158 0.9456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5645 1.8300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3562 2.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1051 1.4081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9867 3.0720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5848 2.5469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7704 1.9170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.0030 0.4875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6388 -0.0149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9891 -0.8726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0535 -0.5088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1507 -0.4991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8068 0.1572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6253 -0.2749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2468 -0.4132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7738 -0.9440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3511 -0.9976 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0885 0.3300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7779 0.3628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0151 -1.9355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5595 -3.0720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
20 15 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
26 29 1 0 0 0 0
21 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 18 1 0 0 0 0
36 40 1 0 0 0 0
22 41 1 0 0 0 0
41 40 1 0 0 0 0
42 43 1 0 0 0 0
8 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 OCH3
M SBV 1 47 -0.8063 0.4655
S SKP 5
ID FL5FDAGS0003
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES OC(C(O)1)C(COC(O5)C(C(C(O)C(C)5)O)O)OC(Oc(c2)cc(O)c(C(=O)3)c2OC(c(c4)ccc(O)c4)=C(OC)3)C(O)1
M END