Mol:FL5FCCGA0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.7548 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7548 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7548 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7548 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1985 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1985 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6422 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6422 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6422 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6422 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1985 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1985 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0859 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0859 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5296 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5296 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5296 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5296 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0859 0.0833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0859 0.0833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0859 -1.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0859 -1.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8291 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8291 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2622 -0.1204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2622 -0.1204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3048 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3048 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3048 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3048 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2622 1.1890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2622 1.1890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8291 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8291 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1985 -1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1985 -1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9116 1.2120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9116 1.2120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9663 -1.2897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9663 -1.2897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2622 1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2622 1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0439 -0.9296 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.0439 -0.9296 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.2569 -1.4506 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.2569 -1.4506 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.3216 -1.2853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3216 -1.2853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9036 -1.4506 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9036 -1.4506 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.2044 -0.9296 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.2044 -0.9296 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.6259 -1.0949 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.6259 -1.0949 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.5148 -1.0793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5148 -1.0793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6259 -0.7120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6259 -0.7120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0238 -0.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0238 -0.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8419 -0.0462 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.8419 -0.0462 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.2436 -0.5764 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2436 -0.5764 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.8220 -0.3515 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.8220 -0.3515 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.5258 -0.4235 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.5258 -0.4235 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.9746 0.0602 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.9746 0.0602 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.4685 -0.2069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4685 -0.2069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6173 -0.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6173 -0.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7744 -0.9974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7744 -0.9974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1632 -0.5942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1632 -0.5942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1120 0.3809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1120 0.3809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6120 1.2469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6120 1.2469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1032 -1.7429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1032 -1.7429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1032 -2.5679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1032 -2.5679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0814 0.4695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0814 0.4695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6647 1.0528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6647 1.0528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 20 1 0 0 0 0 | + | 23 20 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 1 40 1 0 0 0 0 | + | 1 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 35 44 1 0 0 0 0 | + | 35 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 44 45 | + | M SAL 3 2 44 45 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 48 3.3976 0.063 | + | M SVB 3 48 3.3976 0.063 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 46 1.2565 -1.4239 | + | M SVB 2 46 1.2565 -1.4239 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 44 -4.112 0.3809 | + | M SVB 1 44 -4.112 0.3809 |
| − | S SKP 8 | + | S SKP 8 |
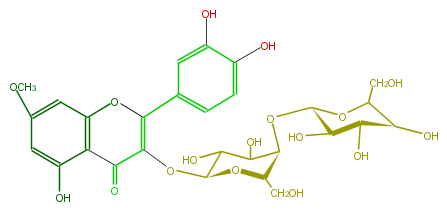
| − | ID FL5FCCGA0002 | + | ID FL5FCCGA0002 |
| − | KNApSAcK_ID C00005510 | + | KNApSAcK_ID C00005510 |
| − | NAME Rhamnetin 3-galactosyl-(1->4)-galactoside | + | NAME Rhamnetin 3-galactosyl-(1->4)-galactoside |
| − | CAS_RN 59920-28-4 | + | CAS_RN 59920-28-4 |
| − | FORMULA C28H32O17 | + | FORMULA C28H32O17 |
| − | EXACTMASS 640.163949598 | + | EXACTMASS 640.163949598 |
| − | AVERAGEMASS 640.54348 | + | AVERAGEMASS 640.54348 |
| − | SMILES c(c(OC)5)c(O)c(C(=O)2)c(c5)OC(=C2O[C@H](O3)C(O)C(O)[C@@H](O[C@@H](O4)[C@H](O)[C@@H]([C@H](O)C4CO)O)[C@H]3CO)c(c1)cc(c(c1)O)O | + | SMILES c(c(OC)5)c(O)c(C(=O)2)c(c5)OC(=C2O[C@H](O3)C(O)C(O)[C@@H](O[C@@H](O4)[C@H](O)[C@@H]([C@H](O)C4CO)O)[C@H]3CO)c(c1)cc(c(c1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-3.7548 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7548 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1985 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6422 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6422 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1985 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0859 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5296 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5296 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0859 0.0833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0859 -1.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8291 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2622 -0.1204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3048 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3048 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2622 1.1890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8291 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1985 -1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9116 1.2120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9663 -1.2897 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2622 1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0439 -0.9296 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.2569 -1.4506 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.3216 -1.2853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9036 -1.4506 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.2044 -0.9296 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.6259 -1.0949 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.5148 -1.0793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6259 -0.7120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0238 -0.2556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8419 -0.0462 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.2436 -0.5764 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8220 -0.3515 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.5258 -0.4235 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.9746 0.0602 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.4685 -0.2069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6173 -0.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7744 -0.9974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1632 -0.5942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1120 0.3809 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6120 1.2469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1032 -1.7429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1032 -2.5679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0814 0.4695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6647 1.0528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 8 1 0 0 0 0
4 3 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 20 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
30 31 1 0 0 0 0
34 39 1 0 0 0 0
1 40 1 0 0 0 0
40 41 1 0 0 0 0
25 42 1 0 0 0 0
42 43 1 0 0 0 0
35 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 44 45
M SBL 3 1 48
M SMT 3 CH2OH
M SVB 3 48 3.3976 0.063
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 CH2OH
M SVB 2 46 1.2565 -1.4239
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 OCH3
M SVB 1 44 -4.112 0.3809
S SKP 8
ID FL5FCCGA0002
KNApSAcK_ID C00005510
NAME Rhamnetin 3-galactosyl-(1->4)-galactoside
CAS_RN 59920-28-4
FORMULA C28H32O17
EXACTMASS 640.163949598
AVERAGEMASS 640.54348
SMILES c(c(OC)5)c(O)c(C(=O)2)c(c5)OC(=C2O[C@H](O3)C(O)C(O)[C@@H](O[C@@H](O4)[C@H](O)[C@@H]([C@H](O)C4CO)O)[C@H]3CO)c(c1)cc(c(c1)O)O
M END