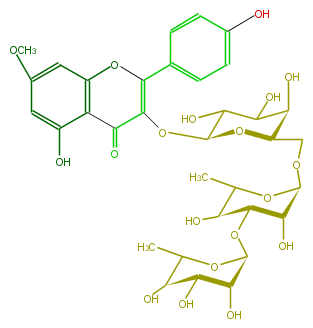
Mol:FL5FCAGS0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.5311 1.6666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5311 1.6666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5311 1.0242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5311 1.0242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9748 0.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9748 0.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4185 1.0242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4185 1.0242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4185 1.6666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4185 1.6666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9748 1.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9748 1.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8622 0.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8622 0.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3059 1.0242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3059 1.0242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3059 1.6666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3059 1.6666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8622 1.9878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8622 1.9878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8622 0.2022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8622 0.2022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2502 1.9876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2502 1.9876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8172 1.6603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8172 1.6603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3842 1.9876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3842 1.9876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3842 2.6423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3842 2.6423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8172 2.9697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8172 2.9697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2502 2.6423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2502 2.6423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0883 0.6353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0883 0.6353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9748 0.0609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9748 0.0609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5391 -0.4785 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.5391 -0.4785 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.2034 -1.0600 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.2034 -1.0600 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.8489 -0.8754 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.8489 -0.8754 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4984 -1.0600 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.4984 -1.0600 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.8342 -0.4785 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.8342 -0.4785 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1886 -0.6629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1886 -0.6629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9327 -0.2577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9327 -0.2577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3135 1.0699 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.3135 1.0699 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.9778 0.4884 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.9778 0.4884 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.6233 0.6730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6233 0.6730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2728 0.4884 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2728 0.4884 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.6086 1.0699 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6086 1.0699 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.9630 0.8855 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.9630 0.8855 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.6900 0.9028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6900 0.9028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2288 1.3459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2288 1.3459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6086 1.7503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6086 1.7503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8883 0.4884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8883 0.4884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7767 -0.0120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7767 -0.0120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7765 -1.1353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7765 -1.1353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5404 -1.4098 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5404 -1.4098 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4984 -1.6264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4984 -1.6264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0181 3.0083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0181 3.0083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4872 -1.8604 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 0.4872 -1.8604 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.1515 -2.4418 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.1515 -2.4418 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.7970 -2.2573 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.7970 -2.2573 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.4465 -2.4418 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.4465 -2.4418 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.7823 -1.8604 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.7823 -1.8604 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.1367 -2.0448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1367 -2.0448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1192 -1.6395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1192 -1.6395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2036 -2.6904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2036 -2.6904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4886 -2.7916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4886 -2.7916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4465 -3.0083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4465 -3.0083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8883 2.2853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8883 2.2853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3883 3.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3883 3.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 8 18 1 0 0 0 0 | + | 8 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 27 33 1 0 0 0 0 | + | 27 33 1 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 24 37 1 0 0 0 0 | + | 24 37 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 21 38 1 0 0 0 0 | + | 21 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 23 40 1 0 0 0 0 | + | 23 40 1 0 0 0 0 |
| − | 28 18 1 0 0 0 0 | + | 28 18 1 0 0 0 0 |
| − | 41 15 1 0 0 0 0 | + | 41 15 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 42 1 0 0 0 0 | + | 47 42 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 39 1 0 0 0 0 | + | 46 39 1 0 0 0 0 |
| − | 1 52 1 0 0 0 0 | + | 1 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 57 -2.8883 2.2853 | + | M SVB 1 57 -2.8883 2.2853 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FCAGS0005 | + | ID FL5FCAGS0005 |
| − | KNApSAcK_ID C00005282 | + | KNApSAcK_ID C00005282 |
| − | NAME Rhamnocitrin 3-rhamninoside | + | NAME Rhamnocitrin 3-rhamninoside |
| − | CAS_RN 39723-40-5 | + | CAS_RN 39723-40-5 |
| − | FORMULA C34H42O19 | + | FORMULA C34H42O19 |
| − | EXACTMASS 754.2320291619999 | + | EXACTMASS 754.2320291619999 |
| − | AVERAGEMASS 754.68588 | + | AVERAGEMASS 754.68588 |
| − | SMILES c(c1C(O6)=C(C(c(c65)c(cc(OC)c5)O)=O)O[C@H](O2)C(C(O)[C@H]([C@@H](CO[C@H](O4)[C@@H](O)[C@H]([C@H](C4C)O)O[C@@H]([C@H]3O)OC([C@H](O)[C@@H]3O)C)2)O)O)cc(cc1)O | + | SMILES c(c1C(O6)=C(C(c(c65)c(cc(OC)c5)O)=O)O[C@H](O2)C(C(O)[C@H]([C@@H](CO[C@H](O4)[C@@H](O)[C@H]([C@H](C4C)O)O[C@@H]([C@H]3O)OC([C@H](O)[C@@H]3O)C)2)O)O)cc(cc1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-2.5311 1.6666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5311 1.0242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9748 0.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4185 1.0242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4185 1.6666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9748 1.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8622 0.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3059 1.0242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3059 1.6666 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8622 1.9878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8622 0.2022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2502 1.9876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8172 1.6603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3842 1.9876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3842 2.6423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8172 2.9697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2502 2.6423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0883 0.6353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9748 0.0609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5391 -0.4785 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.2034 -1.0600 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.8489 -0.8754 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4984 -1.0600 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.8342 -0.4785 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1886 -0.6629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9327 -0.2577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3135 1.0699 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.9778 0.4884 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.6233 0.6730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2728 0.4884 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.6086 1.0699 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.9630 0.8855 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.6900 0.9028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2288 1.3459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6086 1.7503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8883 0.4884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7767 -0.0120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7765 -1.1353 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5404 -1.4098 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4984 -1.6264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0181 3.0083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4872 -1.8604 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.1515 -2.4418 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.7970 -2.2573 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.4465 -2.4418 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.7823 -1.8604 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.1367 -2.0448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1192 -1.6395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2036 -2.6904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4886 -2.7916 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4465 -3.0083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8883 2.2853 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3883 3.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
8 18 1 0 0 0 0
3 19 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 1 0 0 0
29 30 1 1 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 27 1 0 0 0 0
27 33 1 0 0 0 0
32 34 1 0 0 0 0
31 35 1 0 0 0 0
30 36 1 0 0 0 0
24 37 1 0 0 0 0
36 37 1 0 0 0 0
21 38 1 0 0 0 0
22 39 1 0 0 0 0
23 40 1 0 0 0 0
28 18 1 0 0 0 0
41 15 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 39 1 0 0 0 0
1 52 1 0 0 0 0
52 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 57
M SMT 1 OCH3
M SVB 1 57 -2.8883 2.2853
S SKP 8
ID FL5FCAGS0005
KNApSAcK_ID C00005282
NAME Rhamnocitrin 3-rhamninoside
CAS_RN 39723-40-5
FORMULA C34H42O19
EXACTMASS 754.2320291619999
AVERAGEMASS 754.68588
SMILES c(c1C(O6)=C(C(c(c65)c(cc(OC)c5)O)=O)O[C@H](O2)C(C(O)[C@H]([C@@H](CO[C@H](O4)[C@@H](O)[C@H]([C@H](C4C)O)O[C@@H]([C@H]3O)OC([C@H](O)[C@@H]3O)C)2)O)O)cc(cc1)O
M END