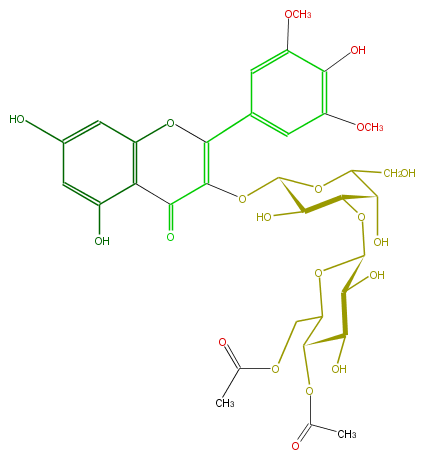
Mol:FL5FAIGA0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 57 0 0 0 0 0 0 0 0999 V2000 | + | 53 57 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.3347 1.1558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3347 1.1558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3347 0.3309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3347 0.3309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6203 -0.0816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6203 -0.0816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9058 0.3309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9058 0.3309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9058 1.1558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9058 1.1558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6203 1.5684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6203 1.5684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1913 -0.0816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1913 -0.0816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4769 0.3309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4769 0.3309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4769 1.1558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4769 1.1558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1913 1.5684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1913 1.5684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1913 -0.7248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1913 -0.7248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4228 1.7271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4228 1.7271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1510 1.3068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1510 1.3068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8792 1.7271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8792 1.7271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8792 2.5679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8792 2.5679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1510 2.9884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1510 2.9884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4228 2.5679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4228 2.5679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1881 1.6486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1881 1.6486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6203 -0.8043 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6203 -0.8043 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4978 3.0231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4978 3.0231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9659 0.4485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9659 0.4485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4818 -0.2326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4818 -0.2326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2247 0.0564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2247 0.0564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9416 0.0641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9416 0.0641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4206 0.5851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4206 0.5851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7707 0.2421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7707 0.2421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2517 0.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2517 0.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7487 -0.3181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7487 -0.3181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6312 -0.3296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6312 -0.3296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9428 -0.8139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9428 -0.8139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4690 -2.9177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4690 -2.9177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3025 -2.7025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3025 -2.7025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2627 -1.8512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2627 -1.8512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6788 -1.1163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6788 -1.1163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7647 -1.4389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7647 -1.4389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8454 -2.3262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8454 -2.3262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5927 -3.8009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5927 -3.8009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1182 -3.3678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1182 -3.3678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8627 -1.4708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8627 -1.4708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3247 -2.4334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3247 -2.4334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9093 -3.3617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9093 -3.3617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1772 -3.3617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1772 -3.3617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1397 -2.8125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1397 -2.8125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2160 -4.0428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2160 -4.0428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5927 -4.4950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5927 -4.4950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3090 -4.9000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3090 -4.9000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2313 -4.6661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2313 -4.6661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1327 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1327 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1881 0.4609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1881 0.4609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1657 3.7476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1657 3.7476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7326 4.9000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7326 4.9000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6080 1.4782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6080 1.4782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8923 1.4782 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8923 1.4782 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 19 3 1 0 0 0 0 | + | 19 3 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 27 8 1 0 0 0 0 | + | 27 8 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 37 45 1 0 0 0 0 | + | 37 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 25 48 1 0 0 0 0 | + | 25 48 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 16 50 1 0 0 0 0 | + | 16 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 14 52 1 0 0 0 0 | + | 14 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 48 49 | + | M SAL 1 2 48 49 |
| − | M SBL 1 1 53 | + | M SBL 1 1 53 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 53 -0.7121 0.0318 | + | M SBV 1 53 -0.7121 0.0318 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 50 51 | + | M SAL 2 2 50 51 |
| − | M SBL 2 1 55 | + | M SBL 2 1 55 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 55 -0.0147 -0.7591 | + | M SBV 2 55 -0.0147 -0.7591 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 52 53 | + | M SAL 3 2 52 53 |
| − | M SBL 3 1 57 | + | M SBL 3 1 57 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 57 -0.7288 0.2489 | + | M SBV 3 57 -0.7288 0.2489 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAIGA0003 | + | ID FL5FAIGA0003 |
| − | FORMULA C33H38O20 | + | FORMULA C33H38O20 |
| − | EXACTMASS 754.1956436559999 | + | EXACTMASS 754.1956436559999 |
| − | AVERAGEMASS 754.64282 | + | AVERAGEMASS 754.64282 |
| − | SMILES C(CO)(C4O)OC(C(C(OC(C5O)OC(COC(C)=O)C(OC(C)=O)C(O)5)4)O)OC(C3=O)=C(Oc(c32)cc(O)cc2O)c(c1)cc(OC)c(O)c(OC)1 | + | SMILES C(CO)(C4O)OC(C(C(OC(C5O)OC(COC(C)=O)C(OC(C)=O)C(O)5)4)O)OC(C3=O)=C(Oc(c32)cc(O)cc2O)c(c1)cc(OC)c(O)c(OC)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 57 0 0 0 0 0 0 0 0999 V2000
-3.3347 1.1558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3347 0.3309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6203 -0.0816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9058 0.3309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9058 1.1558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6203 1.5684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1913 -0.0816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4769 0.3309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4769 1.1558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1913 1.5684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1913 -0.7248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4228 1.7271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1510 1.3068 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8792 1.7271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8792 2.5679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1510 2.9884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4228 2.5679 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1881 1.6486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6203 -0.8043 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4978 3.0231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9659 0.4485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4818 -0.2326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2247 0.0564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9416 0.0641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4206 0.5851 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7707 0.2421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2517 0.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7487 -0.3181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6312 -0.3296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9428 -0.8139 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4690 -2.9177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3025 -2.7025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2627 -1.8512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6788 -1.1163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7647 -1.4389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8454 -2.3262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5927 -3.8009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1182 -3.3678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8627 -1.4708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3247 -2.4334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9093 -3.3617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1772 -3.3617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1397 -2.8125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2160 -4.0428 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5927 -4.4950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3090 -4.9000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2313 -4.6661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1327 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1881 0.4609 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1657 3.7476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7326 4.9000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6080 1.4782 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8923 1.4782 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
4 3 1 0 0 0 0
1 18 1 0 0 0 0
19 3 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
27 8 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
36 40 1 0 0 0 0
34 29 1 0 0 0 0
40 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
42 44 1 0 0 0 0
37 45 1 0 0 0 0
45 46 2 0 0 0 0
45 47 1 0 0 0 0
48 49 1 0 0 0 0
25 48 1 0 0 0 0
50 51 1 0 0 0 0
16 50 1 0 0 0 0
52 53 1 0 0 0 0
14 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 48 49
M SBL 1 1 53
M SMT 1 CH2OH
M SBV 1 53 -0.7121 0.0318
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 50 51
M SBL 2 1 55
M SMT 2 OCH3
M SBV 2 55 -0.0147 -0.7591
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 52 53
M SBL 3 1 57
M SMT 3 OCH3
M SBV 3 57 -0.7288 0.2489
S SKP 5
ID FL5FAIGA0003
FORMULA C33H38O20
EXACTMASS 754.1956436559999
AVERAGEMASS 754.64282
SMILES C(CO)(C4O)OC(C(C(OC(C5O)OC(COC(C)=O)C(OC(C)=O)C(O)5)4)O)OC(C3=O)=C(Oc(c32)cc(O)cc2O)c(c1)cc(OC)c(O)c(OC)1
M END