Mol:FL5FAHGS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.6663 -0.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6663 -0.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6663 -0.7791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6663 -0.7791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1100 -1.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1100 -1.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5536 -0.7791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5536 -0.7791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5536 -0.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5536 -0.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1100 0.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1100 0.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9973 -1.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9973 -1.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4410 -0.7791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4410 -0.7791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4410 -0.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4410 -0.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9973 0.1844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9973 0.1844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9973 -1.6012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9973 -1.6012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8849 0.1843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8849 0.1843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3180 -0.1431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3180 -0.1431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2490 0.1843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2490 0.1843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2490 0.8390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2490 0.8390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3180 1.1663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3180 1.1663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8849 0.8390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8849 0.8390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1100 -1.7424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1100 -1.7424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8850 -1.1002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8850 -1.1002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0427 1.2183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0427 1.2183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2772 0.0617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2772 0.0617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8158 -0.1430 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8158 -0.1430 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3756 0.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3756 0.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0802 -0.3663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0802 -0.3663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6482 -0.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6482 -0.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2197 -0.3663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2197 -0.3663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5151 0.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5151 0.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9471 -0.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9471 -0.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8269 -0.0017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8269 -0.0017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1810 0.3881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1810 0.3881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9414 -0.1008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9414 -0.1008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5627 -0.3396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5627 -0.3396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2772 -0.7521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2772 -0.7521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0346 1.7424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0346 1.7424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6799 1.3299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6799 1.3299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 14 22 1 0 0 0 0 | + | 14 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 22 24 1 0 0 0 0 | + | 22 24 1 0 0 0 0 |
| − | 26 32 1 0 0 0 0 | + | 26 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 16 34 1 0 0 0 0 | + | 16 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 32 33 | + | M SAL 1 2 32 33 |
| − | M SBL 1 1 35 | + | M SBL 1 1 35 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 35 -7.4029 4.1387 | + | M SBV 1 35 -7.4029 4.1387 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 34 35 | + | M SAL 2 2 34 35 |
| − | M SBL 2 1 37 | + | M SBL 2 1 37 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 37 -7.4625 4.6881 | + | M SBV 2 37 -7.4625 4.6881 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAHGS0003 | + | ID FL5FAHGS0003 |
| − | KNApSAcK_ID C00005764 | + | KNApSAcK_ID C00005764 |
| − | NAME Laricitrin 5'-glucoside | + | NAME Laricitrin 5'-glucoside |
| − | CAS_RN 123442-26-2 | + | CAS_RN 123442-26-2 |
| − | FORMULA C22H22O13 | + | FORMULA C22H22O13 |
| − | EXACTMASS 494.10604078999995 | + | EXACTMASS 494.10604078999995 |
| − | AVERAGEMASS 494.40228 | + | AVERAGEMASS 494.40228 |
| − | SMILES OC(C1O)C(Oc(c4O)cc(cc4OC)C(=C2O)Oc(c3)c(c(O)cc3O)C2=O)OC(CO)C1O | + | SMILES OC(C1O)C(Oc(c4O)cc(cc4OC)C(=C2O)Oc(c3)c(c(O)cc3O)C2=O)OC(CO)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-3.6663 -0.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6663 -0.7791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1100 -1.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5536 -0.7791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5536 -0.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1100 0.1844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9973 -1.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4410 -0.7791 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4410 -0.1368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9973 0.1844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9973 -1.6012 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8849 0.1843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3180 -0.1431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2490 0.1843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2490 0.8390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3180 1.1663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8849 0.8390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1100 -1.7424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8850 -1.1002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0427 1.2183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2772 0.0617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8158 -0.1430 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3756 0.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0802 -0.3663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6482 -0.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2197 -0.3663 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5151 0.1453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9471 -0.0170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8269 -0.0017 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1810 0.3881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9414 -0.1008 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5627 -0.3396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2772 -0.7521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0346 1.7424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6799 1.3299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
8 19 1 0 0 0 0
20 15 1 0 0 0 0
1 21 1 0 0 0 0
14 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
22 24 1 0 0 0 0
26 32 1 0 0 0 0
32 33 1 0 0 0 0
16 34 1 0 0 0 0
34 35 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 32 33
M SBL 1 1 35
M SMT 1 CH2OH
M SBV 1 35 -7.4029 4.1387
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 34 35
M SBL 2 1 37
M SMT 2 OCH3
M SBV 2 37 -7.4625 4.6881
S SKP 8
ID FL5FAHGS0003
KNApSAcK_ID C00005764
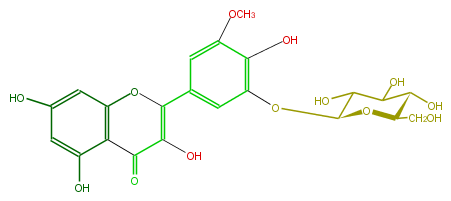
NAME Laricitrin 5'-glucoside
CAS_RN 123442-26-2
FORMULA C22H22O13
EXACTMASS 494.10604078999995
AVERAGEMASS 494.40228
SMILES OC(C1O)C(Oc(c4O)cc(cc4OC)C(=C2O)Oc(c3)c(c(O)cc3O)C2=O)OC(CO)C1O
M END